Team:BGU Israel/Notebook1
From 2014.igem.org
(Difference between revisions)
Stav shamir (Talk | contribs) |
|||
| (5 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
<html> | <html> | ||
| - | + | <section> | |
| - | + | ||
| - | + | ||
<div style="height:12px"> </div> | <div style="height:12px"> </div> | ||
<div style="margin-bottom:10px"> <img src="https://static.igem.org/mediawiki/2014/0/07/BGU14notebookbanner.png" alt=""/></div> | <div style="margin-bottom:10px"> <img src="https://static.igem.org/mediawiki/2014/0/07/BGU14notebookbanner.png" alt=""/></div> | ||
| Line 10: | Line 8: | ||
<div class="clear"></div> | <div class="clear"></div> | ||
| - | <div class="textCont" style="height: | + | <div class="textCont" style="height:150px; background-color:#FFFFFF"> |
| - | <p align="center" style="border: thin solid; border-radius: 8px; background: #5FA6B8; font-style: normal; font-weight: 700; font-size: 24px; font-family: Allan, 'Hoefler Text', 'Liberation Serif', Times, 'Times New Roman', serif; vertical-align: middle;"> Click on the Month or Week to check out what we did <img src="https://static.igem.org/mediawiki/2014/f/f5/BGU14finger-point2.png" width="26px"/></p> | + | <p align="center" style="margin-top:-12px; margin-bottom:5px; border: thin solid; border-radius: 8px; background: #5FA6B8; font-style: normal; font-weight: 700; font-size: 24px; font-family: Allan, 'Hoefler Text', 'Liberation Serif', Times, 'Times New Roman', serif; vertical-align: middle;"> Click on the Month or Week to check out what we did <img src="https://static.igem.org/mediawiki/2014/f/f5/BGU14finger-point2.png" width="26px"/></p> |
<table class="table1" style="width:100%"> | <table class="table1" style="width:100%"> | ||
<tr> | <tr> | ||
| Line 59: | Line 57: | ||
</table> | </table> | ||
</div> | </div> | ||
| - | + | <div id="jump" style="position:relative; bottom:70px"></div> | |
| - | <div id="april" class="textCont" style="height:630px; display: | + | <div id="april" class="textCont" style="height:630px; display:block;"> |
<h3 style="border-bottom:dashed;border-color:#000000">April</h3> | <h3 style="border-bottom:dashed;border-color:#000000">April</h3> | ||
<br> | <br> | ||
| Line 69: | Line 67: | ||
| + | |||
<div class="textContNotebook" style="height:460px; background-image:"https://static.igem.org/mediawiki/2014/2/21/BGU14notebook.png""> | <div class="textContNotebook" style="height:460px; background-image:"https://static.igem.org/mediawiki/2014/2/21/BGU14notebook.png""> | ||
<div class="col2" style=" width:400px;"> | <div class="col2" style=" width:400px;"> | ||
<h3>Lab</h3> | <h3>Lab</h3> | ||
| - | <p> | + | <p>Our Ideas:</p> |
| - | </p> | + | <ol> |
| + | <li><span dir="LTR"> </span>Exocytosis of proteins by E.Coli for treating diseases- especially Diabetes.</li> | ||
| + | <li><span dir="LTR"> </span>Citrus greening- bacterial disease of plants</li> | ||
| + | <li><span dir="LTR"> </span>Skin cancer</li> | ||
| + | <li><span dir="LTR"> </span>Early detection of Salmonella- preventing food poisoning</li> | ||
| + | <li><span dir="LTR"> </span>Modified yeast for longer shelf life of bread </li> | ||
| + | <li><span dir="LTR"> </span>Malaria- one of the largest death causes </li> | ||
| + | <li><span dir="LTR"> </span>Bacterial producing of insulin in the intestine</li> | ||
| + | <li><span dir="LTR"> </span>Methicillin-resistant staphylococcus aureus (MRSA)</li> | ||
| + | </ol> | ||
| + | <p>MALARIA it is!-Lets build a project! </p> | ||
| + | <p>Research is going on!- Reading, talking, arguing and asking</p> | ||
| + | |||
</div> | </div> | ||
<div class="col2" style=" width:400px;margin-left:160px" > | <div class="col2" style=" width:400px;margin-left:160px" > | ||
| - | <h3> | + | <h3>General</h3> |
| - | < | + | <ol> |
| - | + | <li>Looking for the team</li> | |
| - | + | <li>Interviewing students for the team- Finalizing team roster after interviewing </li> | |
| + | <li>FIRST MEETING!-Getting to know each other, What is iGEM competition? Thinking about project ideas</li> | ||
| + | |||
| + | <li align="left">Meeting <strong>with Human Practice team leader of 2013</strong> - Learned about the nature of Human Practice and what its stands for, and got a better understanding of their thinking process about Human Practice strategies. | ||
| + | <br> Starting reading and looking for previous iGEM outstanding projects at the Human Practice field. Maybe</li> | ||
| + | </ol> | ||
| - | |||
</div> | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| - | |||
<div id="may" class="textCont" style="height:630px; display:none"> | <div id="may" class="textCont" style="height:630px; display:none"> | ||
<h3 style="border-bottom:dashed;border-color:#000000">May</h3> | <h3 style="border-bottom:dashed;border-color:#000000">May</h3> | ||
| Line 93: | Line 110: | ||
<div align="center" style="float:right" class="prevNext" onClick="show(june)">Next <img src="https://static.igem.org/mediawiki/2014/d/dd/BGU14finger-point.gif" width="26px"/></div> | <div align="center" style="float:right" class="prevNext" onClick="show(june)">Next <img src="https://static.igem.org/mediawiki/2014/d/dd/BGU14finger-point.gif" width="26px"/></div> | ||
<div class="clear"></div> | <div class="clear"></div> | ||
| - | <div class="textContNotebook" style="height:460px; background-image:" | + | <div class="textContNotebook" style="height:460px; background-image:background-image:"https://static.igem.org/mediawiki/2014/2/21/BGU14notebook.png""> |
| - | + | <div class="col2" style=" width:400px;"> | |
<h3>Lab</h3> | <h3>Lab</h3> | ||
| - | < | + | <ol> |
| + | <li>Are we in the right direction? A lot of dead ends came up and made as wonder if we made the right choice for our research subject. </li> | ||
| + | <li>Malaria is OUT- Metabolic Syndrome- IN: After the disappointment, we came up with a new idea!</li> | ||
| + | <li>Talking about our reading conclusions</li> | ||
| + | <li>Genetic Engineering for non-biologist: Introduction of the world of synthetic biology to our non- biologist team members, with a simple words</li> | ||
| + | <li>Research around the clock!- Reading a lot, looking after experts for consulting </li> | ||
| + | </li> | ||
| + | </ol> | ||
</div> | </div> | ||
| - | + | <div class="col2" style="width:400px;margin-left:160px"> | |
| - | + | <h3>Human Practice</h3> | |
| - | <p>Meeting with the chairman of the BGU Students Union – <strong>Mr. Avi Ben Hillel </strong>< | + | <p>Meeting with the chairman of the BGU Students Union – <strong>Mr. Avi Ben Hillel </strong><br> |
| - | + | 21/5- Presenting our project concept to the <strong>BGU Board of Governors</strong><br> | |
| - | + | Meeting with <strong>Jill Ben-Dor and David Spivak</strong> from the department of Donor and Associate Affairs for promoting our project.<br> | |
| - | + | Meeting with <strong>Yossi Shavit</strong>, Director of Bengis Center for entrepreneurship & Hi-Tech Management. Planned our panel event on the innovation day 2014.</p> | |
| - | + | <p>We need a LOGO!</p> | |
| + | <p>The 44 annual Board of governors meeting- Presenting our idea, vision and plan for the first time, in front of the board of governors</p> | ||
</div> | </div> | ||
| - | |||
| - | |||
</div> | </div> | ||
| + | </div> | ||
<div id="june" class="textCont" style="height:630px;display:none"> | <div id="june" class="textCont" style="height:630px;display:none"> | ||
| Line 121: | Line 145: | ||
<div class="col2" style=" width:400px;"> | <div class="col2" style=" width:400px;"> | ||
<h3>Lab</h3> | <h3>Lab</h3> | ||
| - | < | + | <ol> |
| + | <li>Getting to know the lab- First time at prof. Smadar Cohen's lab, getting familiar with the equipment, facilities and common method we intend to use in our research</li> | ||
| + | <li>The strategies:<br> | ||
| + | 1. Silencing the mRNA of PTP1b protein <br> | ||
| + | 2. Adiponectin overexpression<br> | ||
| + | 3. PGC1-a modest expression circuit | ||
| + | |||
| + | </li> | ||
| + | </ol> | ||
</div> | </div> | ||
<div class="col2" style=" width:400px;margin-left:160px" > | <div class="col2" style=" width:400px;margin-left:160px" > | ||
<h3>Human Practice</h3> | <h3>Human Practice</h3> | ||
| + | <p>Meeting with Assaf Rudich</p> | ||
<p>18/6/14- Producing an expert panel on <b>innovation day 2014</b>– "The Metabolic Syndrome - Could Synthetic Biology Provide a Breakthrough Solution"?</p> | <p>18/6/14- Producing an expert panel on <b>innovation day 2014</b>– "The Metabolic Syndrome - Could Synthetic Biology Provide a Breakthrough Solution"?</p> | ||
<p>Meeting with Deans of Our faculties- Dean of Natural Sciences Faculty, <b>Prof. Jiwchar Ganor</b>; Dean of Engineering Sciences Faculty, <b>Prof. Joseph Kost</b>; and Dean of Humanities and Social Sciences, <b>Prof. David Newman.</b></p> | <p>Meeting with Deans of Our faculties- Dean of Natural Sciences Faculty, <b>Prof. Jiwchar Ganor</b>; Dean of Engineering Sciences Faculty, <b>Prof. Joseph Kost</b>; and Dean of Humanities and Social Sciences, <b>Prof. David Newman.</b></p> | ||
| - | + | <p>Meeting with <b>Dr. Ariel B</b>. Lindner from Paris Descartes University – Understood the meaning of Human Practice more deeply, shaped our HP strategies according to his notes.</p> | |
| + | <p>Innovation day- "The Metabolic syndrome, Diabetes and obesity"</p> | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 143: | Line 177: | ||
<div class="col2" style=" width:400px;"> | <div class="col2" style=" width:400px;"> | ||
<h3>Lab</h3> | <h3>Lab</h3> | ||
| - | < | + | <ol> |
| + | <li>Liquid medium transfering</li> | ||
| + | <li>mRuby DNA extraction- miniprep protocol</li> | ||
| + | </ol> | ||
</div> | </div> | ||
<div class="col2" style=" width:400px;margin-left:160px" > | <div class="col2" style=" width:400px;margin-left:160px" > | ||
| Line 167: | Line 204: | ||
<p><b>Intelligent Medication</b> | <p><b>Intelligent Medication</b> | ||
<p><u>Propagation of pcDNA3-mRuby2</u></p> | <p><u>Propagation of pcDNA3-mRuby2</u></p> | ||
| - | <p>Starters were taken from a pcDNA3-mRuby2 bacterial stab (acquired form Addgene) and incubated in LB containing 100 µg/ml ampicillin for 24 hours at | + | <p>Starters were taken from a pcDNA3-mRuby2 bacterial stab (acquired form Addgene) and incubated in LB containing 100 µg/ml ampicillin for 24 hours at 37?c. |
Plasmid DNA was extracted from using a miniprep kit.</p> | Plasmid DNA was extracted from using a miniprep kit.</p> | ||
</p> | </p> | ||
| Line 210: | Line 247: | ||
<p>Goal: A preliminary experiment to validate we can visualize the co-expression of both mRuby2 and eGFP in CT26 eGFP cell line using confocal microscopy. | <p>Goal: A preliminary experiment to validate we can visualize the co-expression of both mRuby2 and eGFP in CT26 eGFP cell line using confocal microscopy. | ||
Method: | Method: | ||
| - | 20,000 cells were plated in each well of a chamber slide and incubated for 24 hours at | + | 20,000 cells were plated in each well of a chamber slide and incubated for 24 hours at 37?c. |
| - | 4 wells were transfected with pcDNA3-mRuby2 using Lipofectamine 2000 (protocol), and incubated for 48 hours at | + | 4 wells were transfected with pcDNA3-mRuby2 using Lipofectamine 2000 (protocol), and incubated for 48 hours at 37?c. The other 4 wells were mock transfected with Lipofectamine 2000 and incubated in the same conditions for negative control. |
After 48 hours, the transfected cells were viewed at the confocal microscope. | After 48 hours, the transfected cells were viewed at the confocal microscope. | ||
eGFP: excitation - 488 nm, emission - 509 nm | eGFP: excitation - 488 nm, emission - 509 nm | ||
mRuby2: excitation - 559 nm, emission - 600 nm </p> | mRuby2: excitation - 559 nm, emission - 600 nm </p> | ||
<p><u>Results:</u></p> | <p><u>Results:</u></p> | ||
| - | <p><img src="https://static.igem.org/mediawiki/2014/6/6a/BGU14notefig1.png" style="height: | + | <p><img src="https://static.igem.org/mediawiki/2014/6/6a/BGU14notefig1.png" style="height:150px"/></p> |
<p>Co-expression of mRuby2 and eGFP. All three pictures show the same cells. Picture A shows only eGFP expressing cells, picture B shows only mRuby2 expressing cells, and picture C show all cells. </p> | <p>Co-expression of mRuby2 and eGFP. All three pictures show the same cells. Picture A shows only eGFP expressing cells, picture B shows only mRuby2 expressing cells, and picture C show all cells. </p> | ||
| Line 297: | Line 334: | ||
<p align="center"><img src="https://static.igem.org/mediawiki/2014/9/9e/BGU14notefig2.png" style="height:150px"/></p> | <p align="center"><img src="https://static.igem.org/mediawiki/2014/9/9e/BGU14notefig2.png" style="height:150px"/></p> | ||
<p>We conducted the experiment both in 24 well plate (for flow cytometry) and in a chamber slide (for confocal microscopy).<br> | <p>We conducted the experiment both in 24 well plate (for flow cytometry) and in a chamber slide (for confocal microscopy).<br> | ||
| - | The cells were plated on the appropriate plates for the different tests (100,000 cells per well in the 24 well plate and 15,000 cells per well in the chamber slide) and incubated for 24 hours at | + | The cells were plated on the appropriate plates for the different tests (100,000 cells per well in the 24 well plate and 15,000 cells per well in the chamber slide) and incubated for 24 hours at 37?c.<br> |
After 24 hours, the incubated cells were transfected (according to the different treatments detailed above) with one or more of the following: </p> | After 24 hours, the incubated cells were transfected (according to the different treatments detailed above) with one or more of the following: </p> | ||
<ul style="list-style:disc"> | <ul style="list-style:disc"> | ||
| Line 303: | Line 340: | ||
<li>scRNA – both detection part A + hair pin part B - 100 pmol for each of the parts (A+B)</li> | <li>scRNA – both detection part A + hair pin part B - 100 pmol for each of the parts (A+B)</li> | ||
</ul> | </ul> | ||
| - | <p>Transfection was done by lipofectamine (protocol), and then the cells were incubated for 48 hours at | + | <p>Transfection was done by lipofectamine (protocol), and then the cells were incubated for 48 hours at 37?c.<br> |
Preparations for image stream were done by the general protocol (protocol). | Preparations for image stream were done by the general protocol (protocol). | ||
</p> | </p> | ||
| Line 436: | Line 473: | ||
DNA parts and backbone were digested with restriction enzymes: Pst1 and EcoR1<br> | DNA parts and backbone were digested with restriction enzymes: Pst1 and EcoR1<br> | ||
The transformation was done according to the <a href="https://www.dropbox.com/s/p6fau982fhzeihe/08-Transformation%20to%20competent%20bactiria.docx?dl=0">general transformation protocol</a>.<br> | The transformation was done according to the <a href="https://www.dropbox.com/s/p6fau982fhzeihe/08-Transformation%20to%20competent%20bactiria.docx?dl=0">general transformation protocol</a>.<br> | ||
| - | At first, heat-shock transformation to chemically competent | + | At first, heat-shock transformation to chemically competent BH5? bacteria with the following DNA parts (separately): </p> |
<ul style="list-style:disc"> | <ul style="list-style:disc"> | ||
<li><span dir="LTR"> </span>pUC 57 Adiponectin</li> | <li><span dir="LTR"> </span>pUC 57 Adiponectin</li> | ||
| Line 444: | Line 481: | ||
<li><span dir="LTR"> </span>pcDNA3.1 UCP1</li> | <li><span dir="LTR"> </span>pcDNA3.1 UCP1</li> | ||
</ul> | </ul> | ||
| - | <p>The transformed bacteria were incubated for 24 hr at | + | <p>The transformed bacteria were incubated for 24 hr at 37?c.<br> |
| - | After 24 hr incubation, colonies from each plate were transferred to liquid medium and incubated for 24 hr at | + | After 24 hr incubation, colonies from each plate were transferred to liquid medium and incubated for 24 hr at 37?c. <br> |
Plasmid DNA was extracted using a miniprep kit and concentrations were measured using nanodrop. <br> | Plasmid DNA was extracted using a miniprep kit and concentrations were measured using nanodrop. <br> | ||
The plasmids were digested with restriction enzymes Pst1 and EcoR1, following <a href="https://www.dropbox.com/s/bvv0w3rod9dabvu/09-restriction.docx?dl=0">general restriction protocol</a>. <br> | The plasmids were digested with restriction enzymes Pst1 and EcoR1, following <a href="https://www.dropbox.com/s/bvv0w3rod9dabvu/09-restriction.docx?dl=0">general restriction protocol</a>. <br> | ||
| Line 509: | Line 546: | ||
DNA parts and backbone were digested with restriction enzymes: Pst1 and EcoR1<br> | DNA parts and backbone were digested with restriction enzymes: Pst1 and EcoR1<br> | ||
The transformation was done according to the <a class="link1" href="https://www.dropbox.com/s/p6fau982fhzeihe/08-Transformation%20to%20competent%20bactiria.docx?dl=0">general transformation protocol</a>.<br> | The transformation was done according to the <a class="link1" href="https://www.dropbox.com/s/p6fau982fhzeihe/08-Transformation%20to%20competent%20bactiria.docx?dl=0">general transformation protocol</a>.<br> | ||
| - | At first, heat-shock transformation to chemically competent | + | At first, heat-shock transformation to chemically competent BH5? bacteria with the following DNA parts (separately): </p> |
<ul style="list-style:disc"> | <ul style="list-style:disc"> | ||
<li><span dir="LTR"> </span>pUC 57 Adiponectin</li> | <li><span dir="LTR"> </span>pUC 57 Adiponectin</li> | ||
| Line 517: | Line 554: | ||
<li><span dir="LTR"> </span>pcDNA3.1 UCP1</li> | <li><span dir="LTR"> </span>pcDNA3.1 UCP1</li> | ||
</ul> | </ul> | ||
| - | <p>The transformed bacteria were incubated for 24 hr at | + | <p>The transformed bacteria were incubated for 24 hr at 37?c.<br> |
| - | After 24 hr incubation, colonies from each plate were transferred to liquid medium and incubated for 24 hr at | + | After 24 hr incubation, colonies from each plate were transferred to liquid medium and incubated for 24 hr at 37?c. <br> |
Plasmid DNA was extracted using a miniprep kit and concentrations were measured using nanodrop. <br> | Plasmid DNA was extracted using a miniprep kit and concentrations were measured using nanodrop. <br> | ||
The plasmids were digested with restriction enzymes Pst1 and EcoR1, following <a class="link1" href="https://www.dropbox.com/s/bvv0w3rod9dabvu/09-restriction.docx?dl=0">general restriction protocol</a>. <br> | The plasmids were digested with restriction enzymes Pst1 and EcoR1, following <a class="link1" href="https://www.dropbox.com/s/bvv0w3rod9dabvu/09-restriction.docx?dl=0">general restriction protocol</a>. <br> | ||
The digested DNA parts were tested by gel electroporation, following <a class="link1" href="https://www.dropbox.com/s/r98k3xv1svh48ys/10-Gel%20Elctrophoresis.docx?dl=0">general gel electrophoresis protocol</a>.<br> | The digested DNA parts were tested by gel electroporation, following <a class="link1" href="https://www.dropbox.com/s/r98k3xv1svh48ys/10-Gel%20Elctrophoresis.docx?dl=0">general gel electrophoresis protocol</a>.<br> | ||
| - | Ligation products were transformed by heat-shock to chemically competent | + | Ligation products were transformed by heat-shock to chemically competent DH5? bacteria, plated on cmp LB agar plates and incubated for 24 hr at 37?c.<br> |
<u>Results: </u><br> | <u>Results: </u><br> | ||
All parts were successfully restricted (unfortunately the picture taken was not in good quality).<u></u><br> | All parts were successfully restricted (unfortunately the picture taken was not in good quality).<u></u><br> | ||
| Line 647: | Line 684: | ||
<u>Results:</u> | <u>Results:</u> | ||
<p><img src="https://static.igem.org/mediawiki/2014/f/f0/BGU14Notefig15.png" style="height:240px"/></p> | <p><img src="https://static.igem.org/mediawiki/2014/f/f0/BGU14Notefig15.png" style="height:240px"/></p> | ||
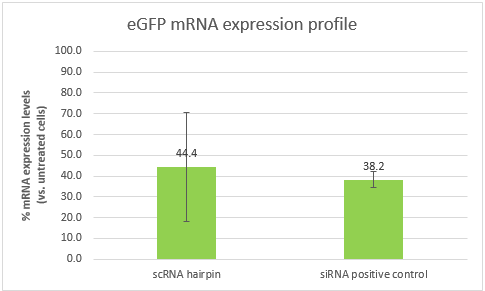
| - | <p>The results were analyzed USING THE | + | <p>The results were analyzed USING THE ??CT method. The cells transfected with our scRNA hairpin contained only 44.4% eGFP mRNA in comparison to untreated cells (with a high standard deviation of 26.1%), while cells transfected with our positive control siRNA contained 38.2% mRNA. </p> |
<u>Conclusions:</u> | <u>Conclusions:</u> | ||
<p>The ability of our scRNA hairpin to perform silencing when it is unbound by the detection part of our construct is of crucial importance. These results show that it can silence a gene of interest, and in a similar efficiency to that of our positive control. However, the high standard deviation, will require us to repeat the experiment. It is also important to note that the efficiency of both samples is affected by the transfection efficiency, which is dependent of the transfection reagent. Optimizing the transfection might lead to better results and higher silencing efficiencies. </p> | <p>The ability of our scRNA hairpin to perform silencing when it is unbound by the detection part of our construct is of crucial importance. These results show that it can silence a gene of interest, and in a similar efficiency to that of our positive control. However, the high standard deviation, will require us to repeat the experiment. It is also important to note that the efficiency of both samples is affected by the transfection efficiency, which is dependent of the transfection reagent. Optimizing the transfection might lead to better results and higher silencing efficiencies. </p> | ||
| Line 672: | Line 709: | ||
<em>Goal: Use TMRM (tetramethylrhodamine methyl ester) to assess the ability of UCP1 to collapse membrane potential. Membrane potential-driven accumulation of TMRM within the inner membrane region of healthy functioning mitochondria results in a dramatic increase in TMRM-associated orange fluorescence. When the mitochondrial membrane potential collapses TMRM is dispersed throughout the cell cytosol at a concentration that yields minimal fluorescence upon excitation in the optimal wavelength region.</em><br> | <em>Goal: Use TMRM (tetramethylrhodamine methyl ester) to assess the ability of UCP1 to collapse membrane potential. Membrane potential-driven accumulation of TMRM within the inner membrane region of healthy functioning mitochondria results in a dramatic increase in TMRM-associated orange fluorescence. When the mitochondrial membrane potential collapses TMRM is dispersed throughout the cell cytosol at a concentration that yields minimal fluorescence upon excitation in the optimal wavelength region.</em><br> | ||
<u>Details:</u><br> | <u>Details:</u><br> | ||
| - | HepG2 cells were seeded in a 24 well plate (75,000 cells per well) and incubated for 24 hours at | + | HepG2 cells were seeded in a 24 well plate (75,000 cells per well) and incubated for 24 hours at 37?c.<br> |
| - | After 24 hours, in 3 of the wells the cells were transfected with pcDNA3.1 UCP1 using Lipofectamine, and incubated for 24 hours at | + | After 24 hours, in 3 of the wells the cells were transfected with pcDNA3.1 UCP1 using Lipofectamine, and incubated for 24 hours at 37?c. <br> |
Then, the cells were dyed with TMRM:</p> | Then, the cells were dyed with TMRM:</p> | ||
<ol> | <ol> | ||
<li><span dir="LTR"> </span>The overnight medium was aspirated.</li> | <li><span dir="LTR"> </span>The overnight medium was aspirated.</li> | ||
<li><span dir="LTR"> </span>Three different concentrations of TMRM 10 µM were added to 4 wells without any treatment: 0 nM, 10 nM, 25 nM, 50 nM, by adding 1 ml of medium DMEM with 0 µl, 1 µl, 2.5 µl, 5 µl of stock solution respectively<span dir="RTL"> </span><span dir="RTL"> </span><span dir="RTL"><span dir="RTL"> </span><span dir="RTL"> </span>.</span></li> | <li><span dir="LTR"> </span>Three different concentrations of TMRM 10 µM were added to 4 wells without any treatment: 0 nM, 10 nM, 25 nM, 50 nM, by adding 1 ml of medium DMEM with 0 µl, 1 µl, 2.5 µl, 5 µl of stock solution respectively<span dir="RTL"> </span><span dir="RTL"> </span><span dir="RTL"><span dir="RTL"> </span><span dir="RTL"> </span>.</span></li> | ||
| - | <li><span dir="LTR"> </span>Incubate for 30 min in | + | <li><span dir="LTR"> </span>Incubate for 30 min in 37?C.</li> |
<li><span dir="LTR"> </span>The medium was replaced to DMEM without phenol red, 0.5 ml in each well.</li> | <li><span dir="LTR"> </span>The medium was replaced to DMEM without phenol red, 0.5 ml in each well.</li> | ||
<li><span dir="LTR"> </span>Same treatment was done with 3 wells of pcDNA3.1 UCP1 transfected HepG2 cells.</li> | <li><span dir="LTR"> </span>Same treatment was done with 3 wells of pcDNA3.1 UCP1 transfected HepG2 cells.</li> | ||
| Line 748: | Line 785: | ||
</section> | </section> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| Line 766: | Line 791: | ||
</html> | </html> | ||
| + | {{Team:BGU_Israel/Footer}} | ||
Latest revision as of 15:36, 17 October 2014


Click on the Month or Week to check out what we did 
| April | May | June | July | August | September | October |
|---|---|---|---|---|---|---|
| Week 2 | Week 6 | Week 10 | ||||
| Week 3 | Week 7 | Week 11 | ||||
| Week 4 | Week 8 | Week 12 | ||||
| Week 1 | Week 5 | Week 9 | Week 13 |
April
 Prev
PrevNext 

Lab
Our Ideas:
- Exocytosis of proteins by E.Coli for treating diseases- especially Diabetes.
- Citrus greening- bacterial disease of plants
- Skin cancer
- Early detection of Salmonella- preventing food poisoning
- Modified yeast for longer shelf life of bread
- Malaria- one of the largest death causes
- Bacterial producing of insulin in the intestine
- Methicillin-resistant staphylococcus aureus (MRSA)
MALARIA it is!-Lets build a project!
Research is going on!- Reading, talking, arguing and asking
General
- Looking for the team
- Interviewing students for the team- Finalizing team roster after interviewing
- FIRST MEETING!-Getting to know each other, What is iGEM competition? Thinking about project ideas
- Meeting with Human Practice team leader of 2013 - Learned about the nature of Human Practice and what its stands for, and got a better understanding of their thinking process about Human Practice strategies.
Starting reading and looking for previous iGEM outstanding projects at the Human Practice field. Maybe
 "
"