Team:Austin Texas/kit
From 2014.igem.org
Jordanmonk (Talk | contribs) m (→Discussion) |
Jordanmonk (Talk | contribs) m |
||
| (154 intermediate revisions not shown) | |||
| Line 10: | Line 10: | ||
<table id="menu" width="100%" cellspacing="0"> | <table id="menu" width="100%" cellspacing="0"> | ||
| - | <tr height=" | + | <tr height="12px"><td></td></tr> |
<tr> | <tr> | ||
<td></td> | <td></td> | ||
| - | |||
| + | |||
| + | <td width="975px" align="left" bgColor="#FFFFFF" cellspacing="0" cellpadding="10px"> | ||
<table width="100%" align="center"> | <table width="100%" align="center"> | ||
| - | |||
| - | |||
| - | |||
| - | <td align="center" onMouseOver="this.bgColor='# | + | <tr height="20px"> |
| - | <a href="https://2014.igem.org/Team:Austin_Texas | + | <td rowspan=3 align="center" onMouseOver="this.bgColor='#000000'" onMouseOut="this.bgColor='#CC6600'" bgColor=#CC6600> |
| + | <a href="https://2014.igem.org/Team:Austin_Texas"style="text-decoration:none;color:#1C140D"><font color=#FFFFFF> | ||
| + | UT Austin<br>HOME </font></a> </td> | ||
| + | <td colspan=3 align="center" bgColor=#666666> <font color=#FFFFFF>PROJECTS</font></td> | ||
| + | <td colspan=1 align="center" bgColor=#666666> <font color=#FFFFFF>HUMAN PRACTICES</font></td> | ||
| + | <td colspan=2 align="center" bgColor=#666666> <font color=#FFFFFF>OFFICIAL REQUIREMENTS</font></td> | ||
| + | <td rowspan=3 align="center" onMouseOver="this.bgColor='#000000'" onMouseOut="this.bgColor='#CC6600'" bgColor=#CC6600 | ||
| + | ><a href="https://igem.org/Main_Page"> <img src="https://static.igem.org/mediawiki/igem.org/6/60/Igemlogo_300px.png" width="35px"></a></td> | ||
| + | </tr> | ||
| - | |||
| - | |||
| - | < | + | <tr height="20px"> |
| - | + | ||
| - | <td align="center" onMouseOver="this.bgColor='# | + | <td rowspan=2 align="center" onMouseOver="this.bgColor='#000000'" onMouseOut="this.bgColor='#CC6600'" bgColor=#CC6600> |
| - | <a href="https://2014.igem.org/Team:Austin_Texas/ | + | <a href="https://2014.igem.org/Team:Austin_Texas/kit" style="text-decoration:none;color:#1C140D"><font color=#FFFFFF>Expanded Genetic Code<br>Measurement Kit</font></a> </td> |
| - | <td align="center" onMouseOver="this.bgColor='# | + | <td rowspan=2 align="center" onMouseOver="this.bgColor='#000000'" onMouseOut="this.bgColor='#CC6600'" bgColor=#CC6600> |
| - | <a href="https://2014.igem.org/Team:Austin_Texas/ | + | <a href="https://2014.igem.org/Team:Austin_Texas/photocage" style="text-decoration:none;color:#1C140D"><font color=#FFFFFF>Photocaged<br>T7 RNA Polymerase</font></a> </td> |
| - | <td align="center" onMouseOver="this.bgColor='# | + | <td rowspan=2 align="center" onMouseOver="this.bgColor='#000000'" onMouseOut="this.bgColor='#CC6600'" bgColor=#CC6600> |
| - | <a href="https://2014.igem.org/Team:Austin_Texas/ | + | <a href="https://2014.igem.org/Team:Austin_Texas/interlab_study" style="text-decoration:none;color:#1C140D"><font color=#FFFFFF>Interlab<br>Study</font></a></td> |
| - | + | ||
| - | >< | + | |
| - | + | ||
| - | + | ||
| - | < | + | <td rowspan=2 align="center" onMouseOver="this.bgColor='#000000'" onMouseOut="this.bgColor='#CC6600'" bgColor=#CC6600> |
| - | + | <a href="https://2014.igem.org/Team:Austin_Texas/human_practices" style="text-decoration:none;color:#1C140D"><font color=#FFFFFF>Caffeinated coli<br>@SXSW Create</font></a></td> | |
| - | < | + | |
| - | </ | + | |
| - | </ | + | |
| - | < | + | <td align="center" onMouseOver="this.bgColor='#000000'" onMouseOut="this.bgColor='#CC6600'" bgColor=#CC6600> |
| - | < | + | <a href="https://2014.igem.org/Team:Austin_Texas/team" style="text-decoration:none;color:#1C140D"><font color=#FFFFFF>Team</font></a> </td> |
| - | < | + | |
| - | <td align="center" | + | <td align="center" onMouseOver="this.bgColor='#000000'" onMouseOut="this.bgColor='#CC6600'" bgColor=#CC6600> |
| - | onMouseOver="this.bgColor='# | + | <a href="https://2014.igem.org/Team:Austin_Texas/parts" style="text-decoration:none;color:#1C140D"><font color=#FFFFFF>Parts</font></a> </td> |
| - | <a href="https://2014.igem.org/Team:Austin_Texas/ | + | |
| - | < | + | <tr height="20px"> |
| - | <td align="center" | + | |
| - | onMouseOver="this.bgColor='# | + | <td align="center" onMouseOver="this.bgColor='#000000'" onMouseOut="this.bgColor='#CC6600'" bgColor=#CC6600> |
| - | <a href="https://2014.igem.org/Team:Austin_Texas/ | + | <a href="https://2014.igem.org/Team:Austin_Texas/safety" style="text-decoration:none;color:#1C140D"><font color=#FFFFFF>Safety</font></a> </td> |
| - | </td> | + | |
| - | <td align="center" | + | <td align="center" onMouseOver="this.bgColor='#000000'" onMouseOut="this.bgColor='#CC6600'" bgColor=#CC6600> |
| - | onMouseOver="this.bgColor='# | + | <a href="https://2014.igem.org/Team:Austin_Texas/medal_requirements" style="text-decoration:none;color:#1C140D"><font color=#FFFFFF>Medals</font></a> </td> |
| - | <a href="https://2014.igem.org/Team:Austin_Texas/ | + | |
| + | </tr> | ||
| + | <tr height="34px"><td colspan=9 align="center" bgColor=#000000><font color=#FFFFFF size=+2>Expanded Genetic Code Measurement Kit</font> | ||
</td> | </td> | ||
| - | |||
</tr> | </tr> | ||
| + | |||
</table> | </table> | ||
| Line 74: | Line 72: | ||
</html> | </html> | ||
<!-- WIKI CONTENT BEGINS --> | <!-- WIKI CONTENT BEGINS --> | ||
| + | [[file: Austin_Texas_Measurement_Kit_Card.png|right]] | ||
| + | __TOC__ | ||
| + | <div align="justify"> | ||
| + | <h1>Kit Introduction</h1> | ||
| - | + | In recent years the ability to expand the genetic code has been made possible by re-coding the amber stop codon, UAG, via the use of modified tRNA synthetase/tRNA pairs. These synthetase/tRNA pairs act together to charge a modified tRNA with a non-canonical amino acid (ncAA), an amino acid that is not one of the 20 amino acids normally encoded by a codon. While the library of ncAA synthetase/tRNA pairs continues to grow, the properties of each pairing have yet to be systematically characterized using a standardized methodology. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | There are certain desired properties of a synthetase/tRNA pair, such as high fidelity and high efficiency, that must be characterized. Without knowing these properties, it is impossible to know how effectively a synthetase/tRNA pair will work and the resulting products will remain vague unless specifically analyzed with each use. The 2014 University of Texas at Austin iGEM Team has developed a standardized method that allows for efficient qualitative and quantitative in vivo characterization of ncAA tRNA synthetase/tRNA pairs. This Expanded Genetic Code Measurement Kit is portable, easy-to-use, and can be quickly used with any synthetase/tRNA pair. | |
| - | + | <h2>Motivation</h2> | |
| - | + | [[File:Translation of ncAA 10-16-14.png|300px|thumb|right|<b>Figure 1.</b> The process of charging a ncAA, followed by its incorporation during translation. To do this successfully, a novel tRNA synthetase must be evolved that specifically recognizes the ncAA.]] | |
| - | + | In order to recode UAG, a synthetase must be mutated to effectively "charge" a ncAA onto the corresponding tRNA. Various methods of directed evolution are typically used to modify a synthetase such that it can interact with and charge a specific ncAA (Liu et al. 2010). The ncAA synthetases available have different levels of reported efficiency, and they are not always well characterized. Many of the ncAA systems are not widely (re)used. They are published in single, short articles lacking full documentation, and our own experiences of working with a number of ncAAs indicate that not all ncAA synthetases are created equal. Thus, we created a standard kit designed to characterize the properties of any ncAA synthetase/tRNA pair. '''Our goal is to produce a measurement kit that is cheap, reproducible, easy to use, AND easily portable (i.e. you don't need a lot of advanced equipment).''' | |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | <h1>Background</h1> | |
| - | + | The genetic code is composed of 64 nucleotide triplets (codons) that code for 20 highly conserved amino acids that are essential to all organisms on Earth. While the genetic code is specific, it is also degenerate, meaning that more than one codon can encode for the incorporation of a specific amino acid. For example, there are six serine codons and three stop codons (called amber, ochre, and opal). By recoding one of the redundant codons, the recoded codon can signal for the incorporation of a non-canonical amino acid (ncAA) rather than the codon's original usage. Of the three stop codons, the amber codon is the least abundant and thus, the easiest and most efficient to recode. | |
| + | The Schultz lab was the first to expand the genetic code using a unique synthetase/tRNA pair. This synthetase/tRNA pair originated from <i>M. jannaschii</i> tyrosine RS (Mj-TyrRS) and allowed the cell to incorporate <i>o</i>-methyl-<small>L</small>-tyrosine at the amber codon (Wang et al. 2001). Since then, numerous ncAA synthetase/tRNA pairs have been generated. A total of seven ncAAs were used in addition to tyrosine for our project. [https://2014.igem.org/Team:Austin_Texas/kit#ncAA_Table The full list of amino acids used in this study can be found here]. | ||
| - | The kit consists of a three plasmid system: pStG, pFRYC, and pFRY (Figure | + | It is important to note that complications can arise when the genetic code is recoded. In a normal bacterium, release factor RF1 is responsible for terminating translation when the ribosome reaches an amber stop codon. To expand the potential of ncAAs, it is vital to avoid termination at amber codons. Seeing this need, the Church and Isaacs groups engineered a strain of ''E. coli'' that had all of the amber codons removed from the genome, which allowed them to knock out the RF1 gene (Isaacs et al. 2011). The resulting strain, called "amberless" ''E. coli'', has NO AMBER CODONS. Thus, to better characterize the synthetase/tRNA pairs, we have employed amberless ''E. coli'' as a chassis organism. The advantage of this system is that since there are no amber codons in the genome, we cannot accidentally interfere with any cellular processes. Additionally, since RF1 is knocked out, there is no competition between RF1 and the novel synthetase/tRNA pair. These factors allow us to better assess the fidelity and efficiency of ncAA tRNA synthetase/tRNA pairs. |
| + | |||
| + | <h1>Experimental Design and Method</h1> | ||
| + | |||
| + | <h2>Plasmids</h2> | ||
| + | |||
| + | [[File:Kit Plasmid Alignment 10-14-14.png|570px|thumb|right|<b>Figure 2.</b> | ||
| + | pStG contains the specific ncAA synthetase/tRNA pair being tested. pFRYC and pFRY are the ncAA kit reporter plasmids. pFRYC is the control plasmid that yields GFP and RFP expression regardless of synthetase/tRNA pair or the presence of ncAA. pFRY is a nearly identical plasmid with a single difference, the linker between the GFP and RFP sequences contains an amber codon (star) in place of the tyrosine codon found in pFRYC. Thus, the level of GFP expression for pFRY is directly dependent on the efficiency and fidelity of the synthetase/tRNA pair being tested.]] | ||
| + | |||
| + | The kit consists of a three plasmid system: pStG, pFRYC, and pFRY ('''Figure 2'''). | ||
*'''pStG''' contains the ncAA Synthetase/tRNA pair (St) to be tested as well as a Gentamicin (G) resistance gene. | *'''pStG''' contains the ncAA Synthetase/tRNA pair (St) to be tested as well as a Gentamicin (G) resistance gene. | ||
*'''pFRYC''' is the control plasmid and contains an IPTG-induced reporter system and a kanamycin resistance gene. The reporter system is composed of RFP and sfGFP fused via a linker sequence between the two. | *'''pFRYC''' is the control plasmid and contains an IPTG-induced reporter system and a kanamycin resistance gene. The reporter system is composed of RFP and sfGFP fused via a linker sequence between the two. | ||
| - | *'''pFRY''' is the experimental reporter plasmid and is nearly identical to pFRYC with the exception that its linker sequence contains an amber codon in the middle of the linker, whereas pFRYC contains a tyrosine codon in the same location. | + | *'''pFRY''' is the experimental reporter plasmid and is nearly identical to pFRYC with the exception that its linker sequence contains an amber codon (star in '''Figure 2''') in the middle of the linker, whereas pFRYC contains a tyrosine codon in the same location. |
| + | |||
| + | |||
| - | + | <h2>Using The Kit</h2> | |
To use the kit properly, each culture must have two of the three plasmids from above. There must always be a pStG plasmid as well as either pFRYC or pFRY. However, additional controls can be conducted using only one of the three plasmids. | To use the kit properly, each culture must have two of the three plasmids from above. There must always be a pStG plasmid as well as either pFRYC or pFRY. However, additional controls can be conducted using only one of the three plasmids. | ||
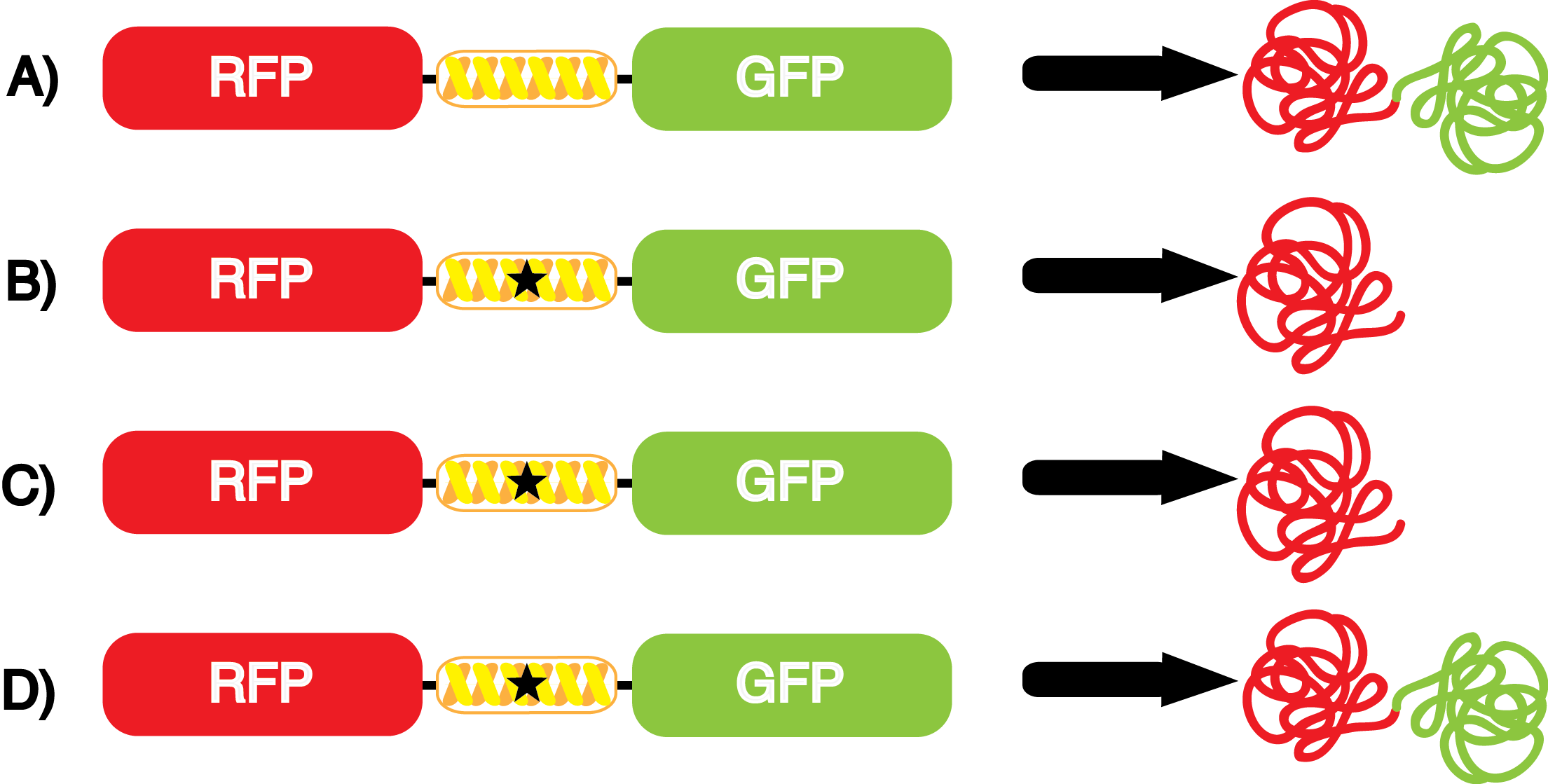
| - | [[File:NcAA kit incorpotation 10-14-14.png|450px|thumb|left|Figure | + | [[File:NcAA kit incorpotation 10-14-14.png|450px|thumb|left|<b>Figure 3.</b> Schematic demonstrating the gene expression of the kit plasmids under different growth conditions in the presence of IPTG.<br> |
A) pFRYC (+/-) pStG, (+/-) ncAA.<br> | A) pFRYC (+/-) pStG, (+/-) ncAA.<br> | ||
B) pFRY (-) pStG, (+/-) ncAA.<br> | B) pFRY (-) pStG, (+/-) ncAA.<br> | ||
C) pFRY (+) pStG, (-) ncAA.<br> | C) pFRY (+) pStG, (-) ncAA.<br> | ||
D) pFRY (+) pStG, (+) ncAA.<br> | D) pFRY (+) pStG, (+) ncAA.<br> | ||
| - | The above results represent the "perfect" tRNA synthetase/tRNA pair: one that has 100% fidelity and 100% efficiency. Actual synthetase/tRNA pairs will occasionally misincorporate an incorrect amino acid, and these pairs will not always be perfectly efficient conducting every step in the process of reading through an amber codon.]] | + | The above results represent the "perfect" tRNA synthetase/tRNA pair: one that has 100% fidelity and 100% efficiency. Actual synthetase/tRNA pairs will occasionally misincorporate an incorrect amino acid, and these pairs will not always be perfectly efficient at conducting every step in the process of reading through an amber codon.]] |
| - | *''' A)''' In a cell containing pStG and pFRYC, the ribosome will translate the RFP reporter, linker, and sfGFP, producing red and green fluorescent proteins that result in visible yellow fluorescence (Figure | + | *''' A)''' In a cell containing pStG and pFRYC, the ribosome will translate the RFP reporter, linker, and sfGFP, producing red and green fluorescent proteins that result in visible yellow fluorescence ('''Figure 3A'''). This happens because the reporter contains no amber codon, and thus does not require a ncAA synthetase/tRNA pair. |
| - | *''' B)''' If a cell contained only pFRY, without pStG, then the ribosome will translate the RFP and terminate at the amber codon on the linker producing a red fluorescence (Figure | + | *''' B)''' If a cell contained only pFRY, without pStG, then the ribosome will translate the RFP and terminate at the amber codon on the linker producing a red fluorescence ('''Figure 3B'''). This occurs because without the pStG, the amber codon is not recoded to allow for ncAA incorporation. |
| - | * '''C)''' In a cell containing pStG and pFRY, there are multiple possible outcomes, depending on what is present in the culture. In the absence of ncAA, the ribosome will translate the RFP and terminate at the amber stop codon on the linker producing a red fluorescence (Figure | + | * '''C)''' In a cell containing pStG and pFRY, there are multiple possible outcomes, depending on what is present in the culture. In the absence of ncAA, the ribosome will translate the RFP and terminate at the amber stop codon on the linker producing a red fluorescence ('''Figure 3C'''). This occurs because while the codon has been recoded, there is no ncAA and without ncAA there can be no incorporation at the amber codon. |
| - | *''' D)''' In a cell containing pStG and pFRY, there are multiple possible outcomes. In the PRESENCE of ncAA, the ribosome will translate the RFP, then it incorporates the ncAA at the amber codon in the linker, and finally it proceeds to translate the downstream sfGFP reporter, producing a visible yellow fluorescence (Figure | + | *''' D)''' In a cell containing pStG and pFRY, there are multiple possible outcomes. In the PRESENCE of ncAA, the ribosome will translate the RFP, then it incorporates the ncAA at the amber codon in the linker, and finally it proceeds to translate the downstream sfGFP reporter, producing a visible yellow fluorescence ('''Figure 3D'''). This occurs because the synthetase found in pStG can covalently attach the ncAA to the tRNA gene present in pStG. This "charged tRNA" is then present during translation, allowing incorporation of the ncAA into the reporter protein. |
| - | However, the above scenarios assume that the synthetase/tRNA pair | + | However, the above scenarios assume that the synthetase/tRNA pair functions perfectly. Some of these systems are non-ideal because they have: |
| - | *Actual synthetase/tRNA pairs sometimes have '''low fidelity''', leading to the incorporation of an incorrect amino acid. | + | *'''LOW FIDELITY:''' Actual synthetase/tRNA pairs sometimes have '''low fidelity''', leading to the incorporation of an incorrect amino acid. This "misincorporation" means that such a synthetase/tRNA pair would not give results that mimic '''Figure 3C''' in the absence of ncAA. Rather, in the absence of ncAA, such a pair would give results similar to '''Figure 3D''', full expression of RFP and sfGFP. High rates of misincorporation are equivalent to '''low fidelity''', and tRNA synthetases with low fidelity are not ideal because they give a mixture of protein products with the normal and new amino acids at the recoded position. |
| - | *Actual synthetase/tRNA pairs can also sometimes have '''low efficiency'''. As these are artificially selected pairs, sometimes they do not function as well as natural synthetase/tRNA pairs. In such a case, the amount of sfGFP translated in Figure | + | *'''LOW EFFICIENCY:''' Actual synthetase/tRNA pairs can also sometimes have '''low efficiency'''. As these are artificially selected pairs, sometimes they do not function as well as natural synthetase/tRNA pairs. In such a case, the amount of sfGFP translated in '''Figure 3D''' might be significantly lower than expected. '''low efficiency''' is also not ideal as it may limit levels of protein expression. |
| + | <h2>Experimental Preparation</h2> | ||
| + | [[File:Alex and Foil-covered incubator.jpg|220px|thumb|right|Protection of light-sensitive ncAAs using a foil-wrapped incubator.]] | ||
| - | + | Each ncAA synthetase/tRNA pair to be tested was cloned into pStG and transformed into pFRYC amberless ''E. coli'' and pFRY amberless ''E. coli''. Other necessary control strains include RFP amberless ''E. coli'' (RFP control), sfGFP amberless ''E. coli'' (GFP control), amberless ''E. coli'' (cell background control), and LB media supplemented with ncAA (media background control). An overnight culture of each strain was grown in LB with the appropriate antibiotics at 37ºC and 225rpm. 10 mL of media with the appropriate antibiotics was inoculated with 100 µL of overnight culture and allowed to grow in the same conditions until the culture density was ~0.2-0.3 OD<sub>600</sub>, or ~3 hours. The 10 mL culture was split between 4 different sterile test-tubes, 2 mL of culture per tube. The conditions of the four test tubes were as follows: | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
*-IPTG,-ncAA | *-IPTG,-ncAA | ||
*-IPTG,+ncAA | *-IPTG,+ncAA | ||
*+IPTG, -ncAA | *+IPTG, -ncAA | ||
*+IPTG, +ncAA | *+IPTG, +ncAA | ||
| - | IPTG | + | If the cultures did not have IPTG or ncAA, an equal volume of sterile deionized water was added in order to keep the volumes between cultures constant. Once the water, the IPTG, and the ncAA were added appropriately, the cultures were allowed to grow to ~0.5 OD<sub>600</sub>. 70 µL of each culture condition and control culture were added to a separate well in clear-bottom black 96-well plate for fluorescence and OD<sub>600</sub> readings using a plate reader. |
| - | + | ||
| + | We accounted for several ncAA-specific concerns. These included: light-sensitivity, oxidation, and interference with fluorescence readings. Certain ncAAs were protected from light due to their light-sensitive molecular structures such as [https://2014.igem.org/Team:Austin_Texas/kit#ncAA_Table ONBY and AzF]. These ncAAs were prepared in a dark room and wrapped in foil. All cultures were grown in a foil-wrapped incubator for consistency. Some ncAAs are more prone to oxidation, specifically <small>L</small>-DOPA. When oxidized, the solution turns black. To prevent oxidation, each ncAA was prepared the day of the test for consistency. By this method, the oxidation of <small>L</small>-DOPA did not occur quickly enough to have an effect on the data. Most ncAA solutions were transparent once prepared with the exception of 3-nitro-<small>L</small>-tyrosine. The yellow-orange tint of 3-nitro-<small>L</small>-tyrosine solution was accounted for by measuring the fluorescence of 1mM 3-nitro-<small>L</small>-tyrosine in media and subtracting any possible background fluorescence from culture fluorescence grown in 1mM 3-nitro-<small>L</small>-tyrosine. | ||
| + | RFP fluorescence was measured using: excitation - 550 nm, emission - 675 nm | ||
| - | + | GFP fluorescence was measured using: excitation - 480 nm, emission - 525 nm | |
| - | + | ||
| + | [[File:10-14-14 big experiment test tubes.png|650px|thumb|left| Preparation for the ncAA Kit Test]] | ||
| + | [[File:RFP-GFP Controls 10-14-15.png|200px|thumb|right| A picture showing culture with or without IPTG. Without IPTG, there is no induction of GFP (left). With IPTG, strong fluorescence is easily seen (right).]] | ||
| Line 183: | Line 182: | ||
| - | |||
| - | + | <h1>Results and Data</h1> | |
| + | The full list of ncAAs and their tRNA synthetase/tRNA pairs can be found in the [https://2014.igem.org/Team:Austin_Texas/kit#ncAA_Table tables at the bottom of this page]. These table include a plethora of information relevant for performing the experiments with each amino acid as well as DNA sequencing data for each synthetase used. | ||
| - | + | <h2>Fidelity of Incorporation</h2> | |
| - | [[File:UT_Austin_2014_Kit_Normalized_GFP_to_RFP_graph.png|thumb|600px|Figure | + | [[File:UT_Austin_2014_Kit_Normalized_GFP_to_RFP_graph.png|thumb|600px|'''Figure 4.''' Graph showing the level of GFP fluorescence relative to RFP fluorescence for each condition. Each pStG plasmid is referred to based on the tRNA synthetase/tRNA pair present in the specific plasmid. Each of these plasmids was then paired with either pFRY or pFRYC and grown in the presence or absence of a specific ncAA. For example, the "3-AminoY-FRYC" and the "3-AminoY-FRY" samples both contain the 3-AminoY synthetase/tRNA pair and both samples were grown in the absence or presence of the ncAA "3-AminoY". Data are presented as the average of three independent cultures. Error bars denote standard deviation.]] |
| - | + | We wanted to test whether the ncAA synthetase/tRNA pairs would incorporate anything besides their amino acid at the amber stop codon (UAG), and that they would in fact incorporate their specific ncAA if it was present. This is essentially the fidelity of each ncAA sythetase/tRNA pair. | |
| + | Fidelity was measured by comparing the production of GFP in cultures containing pStG/pFRY with or without ncAA. In the absence of a ncAA only RFP should be translated, as translation is expected to halt between RFP and GFP at the amber codon on the linker sequence in pFRY. Alternatively, if the corresponding ncAA for pStG is present or if the synthetase/tRNA pair has low fidelity and can misincorporate a different amino acid, translation should continue through the UAG. In this case, RFP and GFP should both be translated. We also tested pStG/pFRYC strains in (+/-) ncAA conditions as a control for what effect the ncAA has on the fluorescence or growth of the cells. These strains should express RFP and GFP in all conditions since pFRYC does not have an amber codon in the linker ('''Figure 2'''). However, if the presence of ncAA affects cell growth or fluorescence activity, we will need these controls to determine the extent of the effect. | ||
| + | To determine the change in GFP fluorescence when the ncAA was present, we first had to calculate how much GFP was expressed relative to the RFP, which would give an upper estimate of how much GFP could theoretically be expressed. We first divided both the GFP and RFP levels by the OD<sub>600</sub> of the culture in order to get the per cell fluorescence levels. We then normalized the GFP fluorescence for one culture to its RFP fluorescence so that we could compare the GFP fluorescence levels between cultures. The normalized GFP values were then compared between cultures grown in the presence of ncAA and cultures grown in the absence of ncAA, which would indicate how the level of GFP fluorescence changes when the ncAA is present. | ||
| - | + | When these values were graphed ('''Figure 4'''), some synthetase/tRNA pairs such as 4-azido-<small>L</small>-phenylalanine (AzF), 3-nitro-<small>L</small>-tyrosine, 3-iodo-<small>L</small>-tyrosine, and ''o''-(2-nitrobenzyl)-<small>L</small>-tyrosine (ONBY) resulted in higher GFP fluorescence in the presence of ncAA than in the absence of ncAA, which suggests that those synthetases only incorporated an amino acid if their specific amino acid was present, meaning that they have a high fidelity. However, the other synthetase/tRNA pairs (3-amino-<small>L</small>-tyrosine, <small>L</small>-DOPA, and 4-cyano-<small>L</small>-phenylalanine (CNF)) did not show a difference in GFP fluorescence normalized to RFP fluorescence dependent on the presence of ncAA, indicating that these pairs incorporated other amino acids at the amber codon when their specific ncAA was not present (and perhaps even when it was), and thus have a low fidelity. | |
| - | + | <h2>Synthetase Efficiency</h2> | |
| - | + | ||
| - | + | ||
| - | + | Another measure of quality for these ncAA synthetase/tRNA pairs is how efficiently they charge their ncAA, ultimately resulting in incorporation of the ncAA in our reporter protein. An inefficient synthetase/tRNA pair will yield incorporation of their ncAA only a fraction of the time, even when their ncAA is present. Our system can be used to measure this level of efficiency, though it does not indicate why a particular synthetase is efficient or inefficient. By comparing the level of GFP fluorescence to the normalized level of RFP fluorescence when the amino acid is present, we can see how efficient the synthetase is. In essence, if the normalized fluorescence of GFP relative to RFP is close to 100% (if the GFP is expressed roughly 100% of the time that RFP is expressed), then the synthetase is very efficient. On the other side, if say the normalized fluorescence of GFP relative to RFP is closer to 10%, then the synthetase would not be very efficient, because even when the ncAA was there, it only incorporated it at the amber codon about 10% of the time. | |
| - | + | For our results ('''Figure 4'''), two synthetase/tRNA pairs stood out as relatively inefficient: 3-nitro-<small>L</small>-tyrosine and ONBY. Both of these synthetase/tRNA pairs showed a significantly smaller normalized GFP to RFP fluorescence when the ncAA was present with the pFRY construct. While both synthetase/tRNA pairs show a significant increase in normalized GFP to RFP fluorescence when the amino acid was present compared to when it was absent, which indicates a high fidelity, the actual GFP fluorescence relative to the RFP fluorescence was only around 50% for 3-nitro-<small>L</small>-tyrosine and 20% for ONBY. These results suggest that these synthetase/tRNA pairs do not always efficiently incorporate their ncAA at an amber stop codon. It is important to note that this could partly result from ncAA-related toxicity, and such an effect was repeatedly observed for ONBY. | |
| - | ''' | + | |
| - | + | ||
| + | <h2>Incorporation Value</h2> | ||
| + | [[File:UT_Austin_2014_Kit_Incorporation_Value_Graph.png|600px|thumb|'''Figure 5.''' Incorporation values for each synthetase/tRNA pair. The dashed line equals an incorporation value of 1. Values greater than 1 when using pFRY indicate high efficiency and fidelity of ncAA incorporation. The GFP:RFP relative fluorescence values in the presence and absence of ncAA (see '''Figure 4''') were used to calculate incorporation values. Values were determined by dividing the GFP:RFP relative fluorescence in the presence of ncAA by the GFP:RFP relative fluorescence in the absence of ncAA. Data are presented as the average of three independent cultures. Error bars denote standard deviation.]] | ||
| - | + | We wanted a way to better summarize the output of the kit with a single measurement value. We decided to calculate an overall '''incorporation value''' as the GFP:RFP relative fluorescence value in the presence of the ncAA divided by the GFP:RFP relative fluorescence value when the ncAA is not included in the culture. Thus, the incorporation value is a single number that measures the relative efficiency and fidelity of a ncAA tRNA synthetase/tRNA pair. A value of 1 indicates no significant dependence of incorporation on the presence of ncAA in the culture. The higher the value, the higher the fidelity and/or efficiency of the synthetase/tRNA pair. It is important to note that ncAA-related effects on growth and translation can also reduce the apparent incorporation value of a synthetase/tRNA pair below 1. | |
| + | '''The synthetase/tRNA pairs that showed the highest incorporation values were AzF, 3-nitrotyrosine, ONBY, and 3-iodotyrosine.''' Among the ncAAs tested, ONBY consistently slowed the growth rate of the culture significantly (as seen by OD<sub>600</sub> readings) suggesting possible toxicity to the cell. However, ncAA incorporation remained relatively high, resulting in a good incorporation value. '''Synthetase/tRNA pairs that showed the lowest levels of ncAA incorporation included 3-amino-<small>L</small>-tyrosine, <small>L</small>-DOPA, and CNF.''' | ||
| - | + | <h1>Discussion</h1> | |
| + | Our ncAA measurement kit was able to successfully compare the fidelity and efficiency of seven different synthetase/tRNA pairs. We were able to confidently say that one synthetase/tRNA pair had a higher fidelity than another. '''We saw clear evidence that three of our tRNA synthetase/tRNA pairs seemed incapable of discriminating between their ncAA and the 20 canonical amino acids present in media: 3-amino-<small>L</small>-tyrosine, <small>L</small>-DOPA, and CNF.''' We were able to test triplicate cultures of seven different ncAAs and tyrosine (eight total synthetase/tRNA pairs) in one day, generating results that immediately told us which ncAAs worked best in our system both in terms of fidelity and efficiency of incorporation. This required no advanced training or access to equipment besides a shaker and a fluorometer. | ||
| - | + | While the measurements may not be perfectly accurate due to the nature of this test, the measurement kit is designed more around ease of use, cost-efficiency, and portability. The ncAA measurement kit can be used to quickly test a synthetase/tRNA pair's quality with minimal effort, as opposed to other more accurate but more intensive or expensive measures such as mass spectrometry. While mass spectrometry can give much more definitive information about a synthetase/tRNA pair, there are many different factors that could make it unreasonable for undergraduate research or high-throughput use. It is a technique that requires a fair amount of skill to do properly, and can be very costly as well. Additionally, the sample needs to be purified from cell culture and prepped for mass spectrometry. | |
| + | For the ncAA measurement kit, all that is necessary is to request from us the kit plasmids (pFRYC and PFRY in separate strains of amberless ''E. coli''), which we are immediately making available to anyone who requests the Expanded Genetic Code Measurement Kit. Once you receive the frozen cultures (or plasmids), transform the synthetase/tRNA pair that you wish to test into the two different cell strains, and run the experiment. Thus, if you receive the measurement kit on Monday, you could have your synthetase/tRNA pairs characterized by Friday. The kit can also be used to test how one or more synthetase/tRNA pairs work in different cells or under different conditions. However, in this case, you may need to retransform the kit plasmids into the different strain. | ||
| - | + | We are optimistic that this measurement kit can help speed up the process of developing a new synthetase/tRNA pairs, as well as characterizing large groups of synthetase/tRNA pairs, as we now have a way to quickly, cheaply, and easily do large scale characterizations. For example, we could now systematically test how changing the expression levels of a synthetase and its tRNA alters the efficiency and fidelity of ncAA incorporation. | |
| - | + | Due to the nature of the Expanded Genetic Code Measurement Kit, <i>in vivo</i> influences will affect certain data and results. This knowledge is very useful by providing a broader and biological perspective of cellular health. In general, the presence of ncAAs slowed the growth of amberless ''E. coli'', sometimes quite significantly. This knowledge, as seen through the GFP:RFP ratio, can be very informative when assessing whether to use a ncAA tRNA synthetase/tRNA pair. | |
| + | <h1>Conclusion</h1> | ||
| + | |||
| + | This year the UT Austin iGEM Team has chosen to take part in the [https://2014.igem.org/Tracks/Measurement Measurement Track] of the iGEM competition. We have presented here the Expanded Genetic Code Measurement Kit for the purpose of quickly characterizing novel ncAA tRNA synthetase/tRNA pairs, which meet the [https://2014.igem.org/Team:Austin_Texas/medal_requirements medal criteria]. Prior to this measurement kit there was not a widely available and easily accessible kit for assessing ncAA tRNA synthetase/tRNA pairs. Although numerous labs have used mass spectrometry, such a technique is not always cheap not widely available. Additionally, we have begun the process of standardization for ncAA synthetase/tRNA pairs by using our kit to simultaneously characterize seven different ncAA synthetase/tRNA pairs as well as a tyrosine RS. '''This method of testing was designed to be easy-to-use, portable, quicker, and more available than other methods of characterization and analysis. We are optimistic that this is a first step in opening the doorway to more iGEM teams and labs using these new chemical building blocks in their projects.''' | ||
| + | |||
| + | Documentation, including sequence information, of the tRNA synthetase/tRNA pairs used can be found [https://2014.igem.org/Team:Austin_Texas/kit#ncAA_Synthetase_Table below]. | ||
| + | |||
| + | Information regarding the ncAAs used can also be found [https://2014.igem.org/Team:Austin_Texas/kit#ncAA_Table below]. | ||
| + | </div> | ||
| + | <h1>ncAA Table</h1> | ||
{| class="wikitable" | {| class="wikitable" | ||
| Line 229: | Line 240: | ||
|- | |- | ||
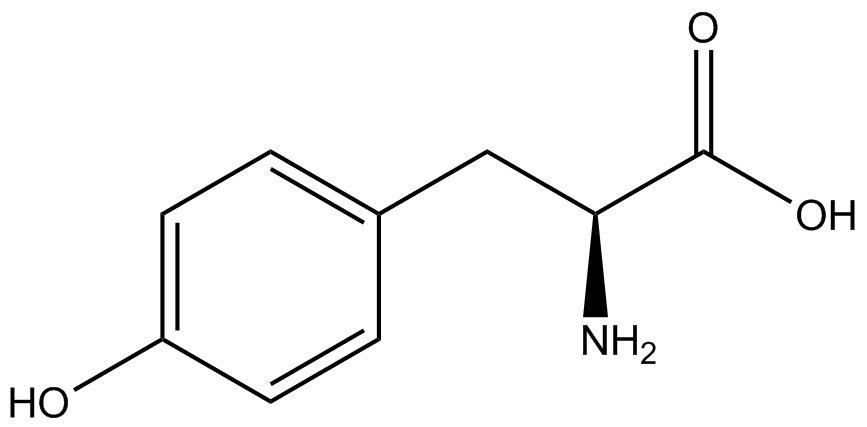
| - | | tyrosine<br> (canonical) | + | | <small>L</small>-tyrosine<br> (canonical) |
| [[Image:Y.png | 150px ]] | | [[Image:Y.png | 150px ]] | ||
| 181.19 | | 181.19 | ||
| Line 236: | Line 247: | ||
|- | |- | ||
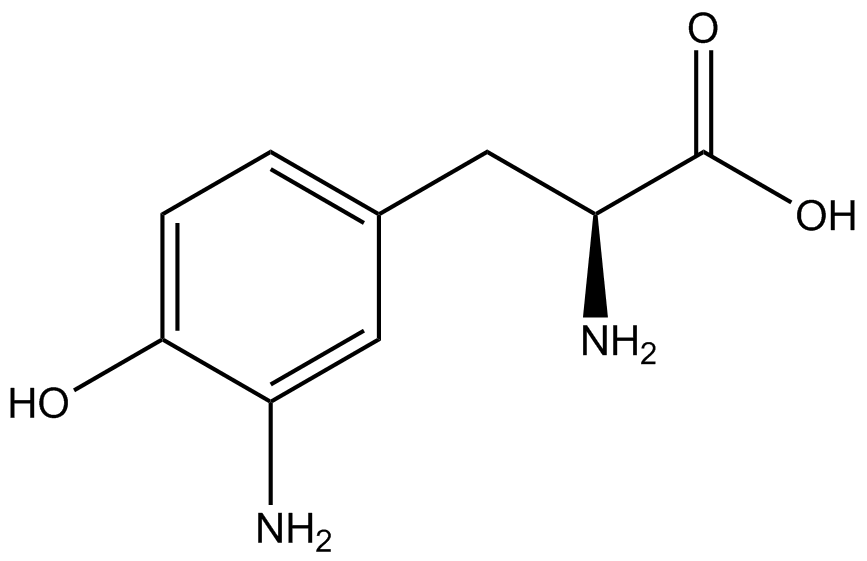
| - | | 3- | + | | 3-amino-<small>L</small>-tyrosine (AminoY) |
| [[Image:AminoY.png | 150px ]] | | [[Image:AminoY.png | 150px ]] | ||
| 287.14 | | 287.14 | ||
| Line 243: | Line 254: | ||
|- | |- | ||
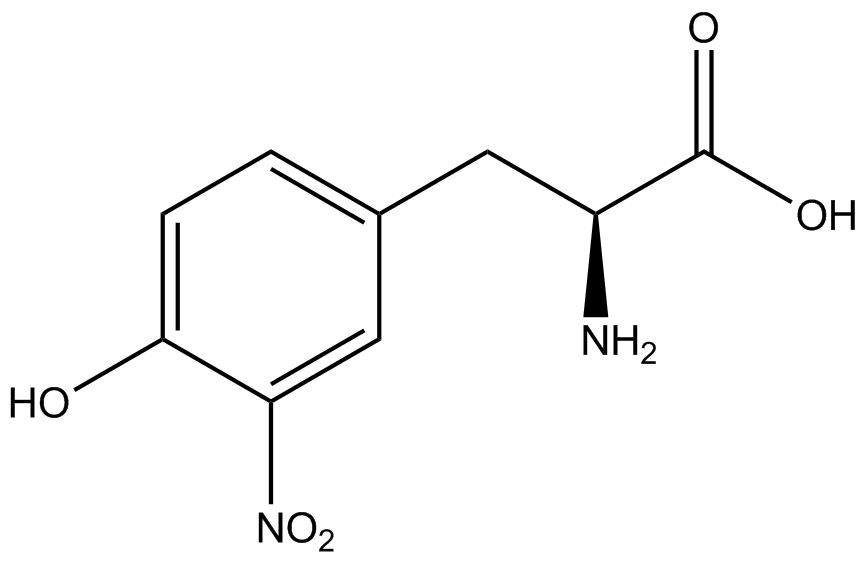
| - | | 3- | + | | 3-nitro-<small>L</small>-tyrosine (NitroY) |
| [[Image:NitroY2.png | 150px ]] | | [[Image:NitroY2.png | 150px ]] | ||
| 226.2 | | 226.2 | ||
| Line 250: | Line 261: | ||
|- | |- | ||
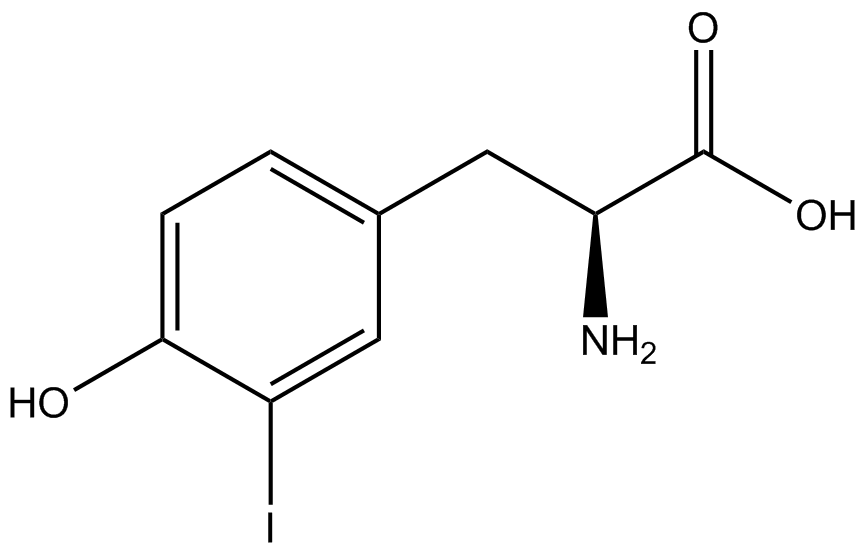
| - | | 3- | + | | 3-iodo-<small>L</small>-tyrosine (IodoY) |
| [[Image:IodoY2.png | 150px ]] | | [[Image:IodoY2.png | 150px ]] | ||
| 307.09 | | 307.09 | ||
| Soluble in water<br> Heat to 70°C and vortex to dissolve | | Soluble in water<br> Heat to 70°C and vortex to dissolve | ||
| - | | Stock Concentration: 10mM <br>(. | + | | Stock Concentration: 10mM <br>(.03g in 10mL H2O)<br> Concentration in Culture: 1mM |
|- | |- | ||
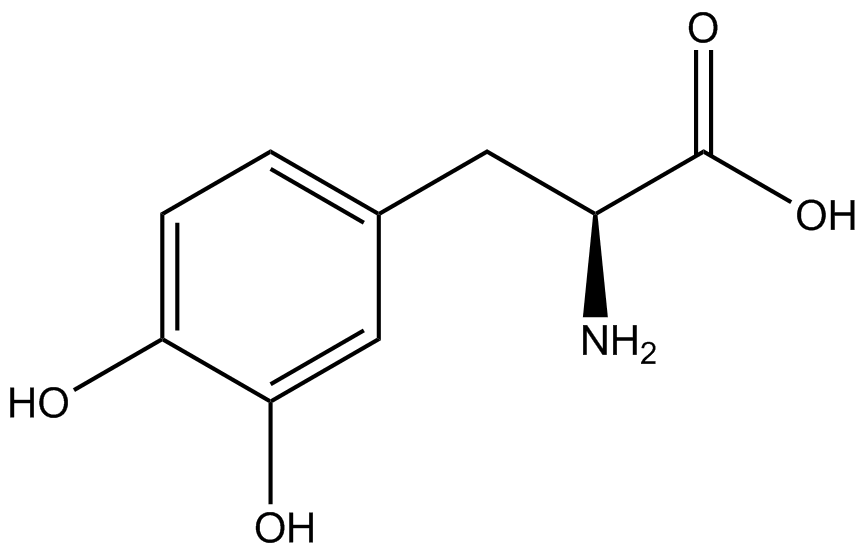
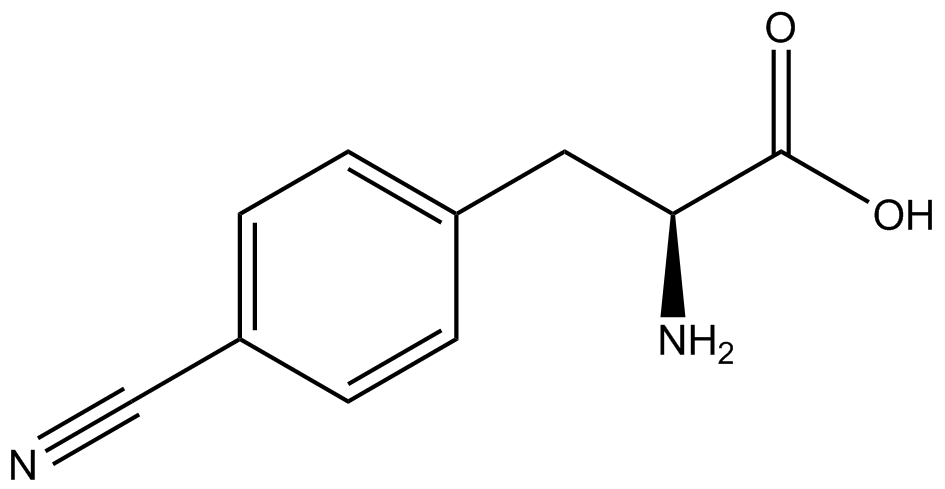
| - | | <small>L</small>- | + | | 3,4-dihydroxy-<small>L</small>-phenylalanine <br>(<small>L</small>-DOPA) |
| [[Image:DOPA2.png | 150px ]] | | [[Image:DOPA2.png | 150px ]] | ||
| 197.1879 | | 197.1879 | ||
| Line 264: | Line 275: | ||
|- | |- | ||
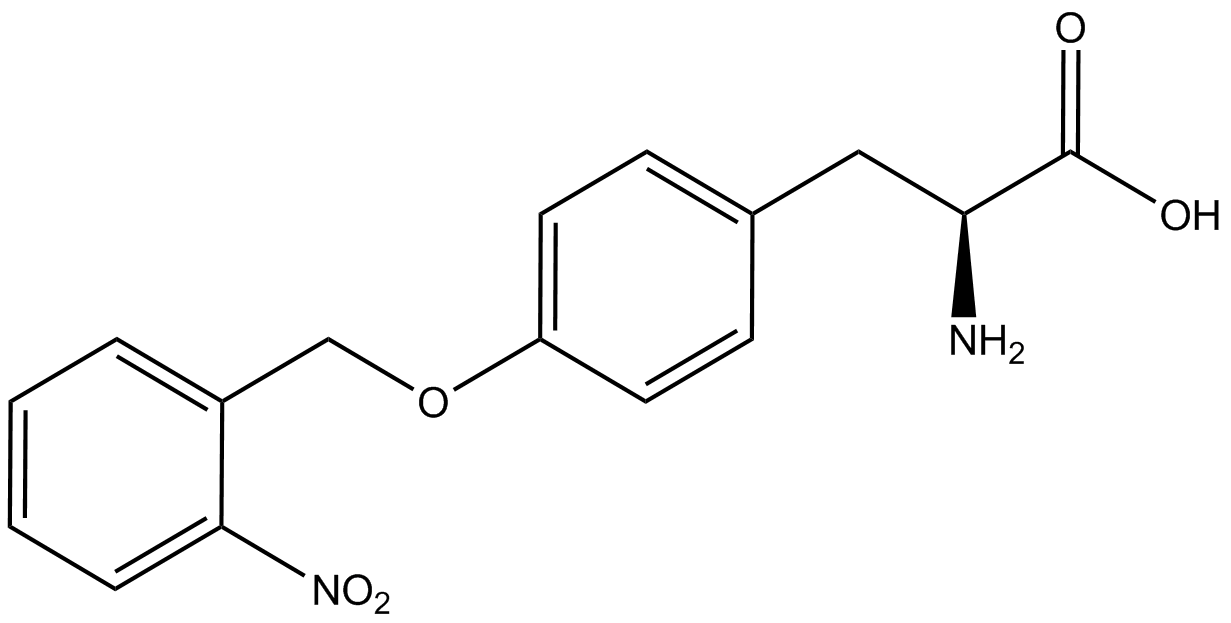
| - | | <i>o</i>-(2-nitrobenzyl)tyrosine <br>(ONBY) | + | | <i>o</i>-(2-nitrobenzyl)-<small>L</small>-tyrosine <br>(ONBY) |
| [[Image:L-ONBY 2.png | 150px ]] | | [[Image:L-ONBY 2.png | 150px ]] | ||
| 316.308 | | 316.308 | ||
| Line 271: | Line 282: | ||
|- | |- | ||
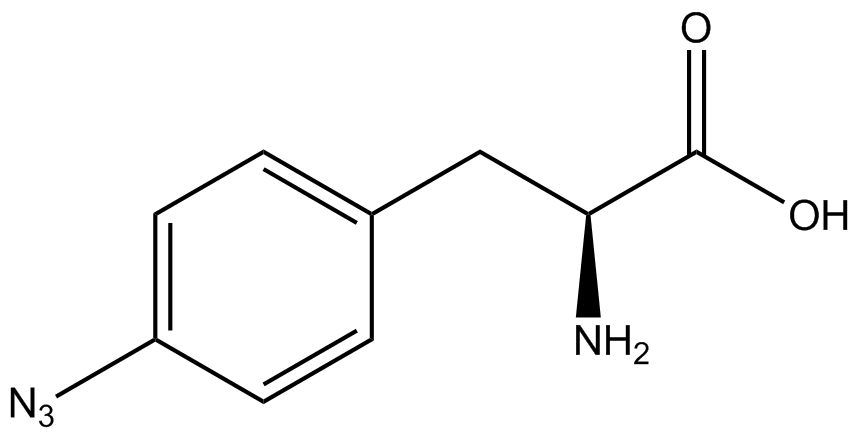
| - | | 4- | + | | 4-azido-<small>L</small>-phenylalanine <br>(AzF) |
| [[Image:AzF2.png | 150px ]] | | [[Image:AzF2.png | 150px ]] | ||
| 206.204 | | 206.204 | ||
| Line 278: | Line 289: | ||
|- | |- | ||
| - | | 4- | + | | 4-cyano-<small>L</small>-phenylalanine <br>(CNF) |
| [[Image:CNF.png | 150px ]] | | [[Image:CNF.png | 150px ]] | ||
| 190.2 | | 190.2 | ||
| Line 286: | Line 297: | ||
|} | |} | ||
| - | + | <h1>ncAA Synthetase Table</h1> | |
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
| Line 296: | Line 307: | ||
! Sequence confirmed? | ! Sequence confirmed? | ||
! Biobrick? | ! Biobrick? | ||
| + | ! Incorporation Value | ||
|- | |- | ||
| Line 303: | Line 315: | ||
| Mj-tRNA Tyr (CUA) | | Mj-tRNA Tyr (CUA) | ||
| Wang et al. 2001. | | Wang et al. 2001. | ||
| - | | [https://2014.igem.org/Team:Austin_Texas/kit/Sequences/TYR_RS | + | | [https://2014.igem.org/Team:Austin_Texas/kit/Sequences/TYR_RS Yes] |
| No | | No | ||
| + | | 1 | ||
|- | |- | ||
| - | | | + | | AminoY RS |
| Mj-TyrRS | | Mj-TyrRS | ||
| Y32Q, L65E, F108G, Q109L, D158S, L162Y | | Y32Q, L65E, F108G, Q109L, D158S, L162Y | ||
| Mj-tRNA Tyr (CUA) | | Mj-tRNA Tyr (CUA) | ||
| Seyedsayamdost et al. 2007. | | Seyedsayamdost et al. 2007. | ||
| - | | [https://2014.igem.org/Team:Austin_Texas/kit/Sequences/3-aminoY_RS | + | | [https://2014.igem.org/Team:Austin_Texas/kit/Sequences/3-aminoY_RS Yes] |
| No | | No | ||
| + | | 1 | ||
|- | |- | ||
| - | | | + | | NitroY RS |
| Mj-TyrRS | | Mj-TyrRS | ||
| Y32R, H70L, Q155M, D158G, I159L, L162H | | Y32R, H70L, Q155M, D158G, I159L, L162H | ||
| Line 323: | Line 337: | ||
| [https://2014.igem.org/Team:Austin_Texas/kit/Sequences/3-nitroY_RS Yes] | | [https://2014.igem.org/Team:Austin_Texas/kit/Sequences/3-nitroY_RS Yes] | ||
| No | | No | ||
| + | | 6 | ||
|- | |- | ||
| - | | | + | | IodoY RS |
| Mj-TyrRS | | Mj-TyrRS | ||
| H70A, D158T, I159S, D286Y | | H70A, D158T, I159S, D286Y | ||
| Mj-tRNA Tyr (CUA) | | Mj-tRNA Tyr (CUA) | ||
| Sakamoto et al. 2009. | | Sakamoto et al. 2009. | ||
| - | | [https://2014.igem.org/Team:Austin_Texas/kit/Sequences/3-iodoY_RS | + | | [https://2014.igem.org/Team:Austin_Texas/kit/Sequences/3-iodoY_RS Yes] |
| [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1416001 BBa_K1416001] | | [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1416001 BBa_K1416001] | ||
| + | | 5 | ||
|- | |- | ||
| Line 339: | Line 355: | ||
| Mj-tRNA Tyr (CUA) | | Mj-tRNA Tyr (CUA) | ||
| Alfonta et al. 2003. | | Alfonta et al. 2003. | ||
| - | | [https://2014.igem.org/Team:Austin_Texas/kit/Sequences/L-DOPA_RS | + | | [https://2014.igem.org/Team:Austin_Texas/kit/Sequences/L-DOPA_RS Yes] |
| [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1178000 BBa_K1178000], 2013 | | [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1178000 BBa_K1178000], 2013 | ||
| + | | 1 | ||
|- | |- | ||
| Line 348: | Line 365: | ||
| Mj-tRNA Tyr (CUA) | | Mj-tRNA Tyr (CUA) | ||
| Deiters et al. 2006. | | Deiters et al. 2006. | ||
| - | | [https://2014.igem.org/Team:Austin_Texas/kit/Sequences/ONBY_RS | + | | [https://2014.igem.org/Team:Austin_Texas/kit/Sequences/ONBY_RS Yes] |
| [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1416000 BBa_K1416000] | | [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1416000 BBa_K1416000] | ||
| + | | 6 | ||
|- | |- | ||
| Line 357: | Line 375: | ||
| Mj-tRNA Tyr (CUA) | | Mj-tRNA Tyr (CUA) | ||
| Chin et al. 2002. | | Chin et al. 2002. | ||
| - | | [https://2014.igem.org/Team:Austin_Texas/kit/Sequences/AzF_RS | + | | [https://2014.igem.org/Team:Austin_Texas/kit/Sequences/AzF_RS Yes] |
| No | | No | ||
| + | | 9 | ||
|- | |- | ||
| Line 366: | Line 385: | ||
| Mj-tRNA Tyr (CUA) | | Mj-tRNA Tyr (CUA) | ||
| Schultz et al. 2006. | | Schultz et al. 2006. | ||
| - | | [https://2014.igem.org/Team:Austin_Texas/kit/Sequences/CNF_RS | + | | [https://2014.igem.org/Team:Austin_Texas/kit/Sequences/CNF_RS Yes] |
| No | | No | ||
| + | | 1 | ||
|} | |} | ||
| - | + | <h1>References</h1> | |
| - | * | + | * Alfonta, L., Zhang, Z., Uryu, S., Loo, J. A., Schultz, P. G. (2003) Site-specific incorporation of a redox-active amino acid into proteins. ''J. Am. Chem. Soc.'' '''125''':14662-14663. |
| - | * Wang, L., | + | * Chin, J. W., Santoro, S. W., Martin, A. B., King, D. S., Wang, L., Schultz, P. G. (2002) Addition of p-azido-<small>L</small>-phenylalanine to the genetic code of ''Escherichia coli''. ''J. Am. Chem. Soc.'' '''1294''':9026-9027. |
| + | * Deiters, A., Groff, D., Ryu, Y., Xie, J., Schultz, P. G. (2006) A genetically encoded photocaged tyrosine. ''Angew. Chem. Int. Ed. Engl.'' '''45''':2728-2731. | ||
| + | *Isaacs, F. J., Carr, P. A., Wang, H. H., Lajoie, M. J., Sterling, B., Kraal, L., Tolonen, A.C., Gianoulis, T.A., Goodman, D.B., Reppas, N.B., Emig, C.J., Bang, D., Hwang, S.J., Jewett, M. C., Jacobson, J. M., Church, G.M. (2011) Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. ''Science'' '''333''':348-353 | ||
| + | *Liu, C. C., Schultz, P. G. (2010) Adding new chemistries to the genetic code. ''Annu. Rev. Biochem.'' '''79''':413-44. | ||
* Neumann, H., Hazen, J. L., Weinstein, J., Mehl, R. A., Chin, J. W. (2008) Genetically encoding protein oxidative damage. ''J. Am. Chem. Soc.'' '''130''': 4028-4033. | * Neumann, H., Hazen, J. L., Weinstein, J., Mehl, R. A., Chin, J. W. (2008) Genetically encoding protein oxidative damage. ''J. Am. Chem. Soc.'' '''130''': 4028-4033. | ||
| - | * Sakamoto, K., Murayama, K., Oki, K., Iraha, F., Kato-Murayama, M., Takahashi, M., Ohtake, K., Kobayashi, T., Kuramitsu, S., Shirouzu, M., Yokoyama, S. (2009) Genetic encoding of 3-iodo-L-tyrosine in Escherichia coli for single-wavelength anomalous dispersion phasing in protein crystallography. ''Structure''. '''17''': 335-344 | + | * Sakamoto, K., Murayama, K., Oki, K., Iraha, F., Kato-Murayama, M., Takahashi, M., Ohtake, K., Kobayashi, T., Kuramitsu, S., Shirouzu, M., Yokoyama, S. (2009) Genetic encoding of 3-iodo-<small>L</small>-tyrosine in ''Escherichia coli'' for single-wavelength anomalous dispersion phasing in protein crystallography. ''Structure''. '''17''': 335-344. |
| - | + | ||
| - | + | ||
| - | + | ||
* Schultz, K. C., Supekova, L., Ryu, Y., Xie, J., Perera, R., Schultz, P. G. (2006) A genetically encoded infrared probe. ''J. Am. Chem. Soc.'' '''129''':13984-13985. | * Schultz, K. C., Supekova, L., Ryu, Y., Xie, J., Perera, R., Schultz, P. G. (2006) A genetically encoded infrared probe. ''J. Am. Chem. Soc.'' '''129''':13984-13985. | ||
| - | + | * Seyedsayamdost, M. R., Xie, J., Chan, C. T. Y., Schultz, P. G., Stubbe, J. (2007) Site-specific insertion of 3-aminotyrosine into subunit α-2 of ''E. coli'' ribonucleotide reductase: Direct evidence for involvement of Y<sub>730</sub> and Y<sub>731</sub> in radical propagation. ''J. Am. Chem. Soc.'' '''129''': 15060–15071. | |
| - | + | * Wang, L., Brock, A., Herberich, B., Schultz, P. G. (2001) Expanding the genetic code of ''Escherichia coli''. ''Science'' '''292''': 498–500. | |
<!-- WIKI CONTENT ENDS --> | <!-- WIKI CONTENT ENDS --> | ||
Latest revision as of 23:54, 18 March 2015
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"