Team:NCTU Formosa/results
From 2014.igem.org
(→PBAN Peptide SDS Protein Electrophoresis) |
(→Magic Power of Our Pyramidal Device) |
||
| (425 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:NCTU Formosa/source/head}} | {{:Team:NCTU Formosa/source/head}} | ||
{{:Team:NCTU Formosa/source/header-results}} | {{:Team:NCTU Formosa/source/header-results}} | ||
| - | {{:Team:NCTU Formosa/source/ | + | {{:Team:NCTU Formosa/source/header0}} |
{{:Team:NCTU Formosa/source/cover-results}} | {{:Team:NCTU Formosa/source/cover-results}} | ||
{{:Team:NCTU Formosa/source/card}} | {{:Team:NCTU Formosa/source/card}} | ||
| Line 10: | Line 10: | ||
{{:Team:NCTU Formosa/source/pyramidal device}} | {{:Team:NCTU Formosa/source/pyramidal device}} | ||
| - | <p> | + | <p> We develop a powerful and specific insect attractive device which combine the use of blue light and PBAN. The video shows that '''our device successfully and effectively attract more insects'''. In this video: </p> |
| - | [ | + | <p> 1. We used our pyramidal device, biobrick part [http://parts.igem.org/Part:BBa_K1415105 BBa_K1415105: Pcons+RBS+PBAN(''Spodoptera litura'')] and its target insect ''Spodoptera litura'' inside an arcrylic chamber for this expiriment. </p> |
| + | <p> 2. The female moth is fed with PBAN in a small beaker covering with plastic wrap. Soon, the female flaps its wings frantically, a sign of sexual stimulation. From this point on, it starts to release pheromones. </p> | ||
| + | <p> 3. We transferred the beaker into our device then placed the device in an acrylic chamber(moth box) with approximately 150 male moths inside to conduct the test. We kept the chamber dark to ensure that blue light would be the only light source inside. We recorded the number of insects entering the device per hour for eleven hours.</p> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | <p> Fig.1-1 shows the '''entering times''' of moths in different conditions. Entering times is defined as the times that moths get into the device. In this experiment, the moth might get out of our device, since we were only using a simple model device without trap, such as nets for keeping the moths. However, we can still clearly see the magic power of our pyramidal device by using blue light with PBAN treatment, which attracted more moths to get into the device than [https://www.youtube.com/watch?v=IXGEvhqHs-c the other experiment (only blue light treatment)]. In addition, we can even observe a unique periodicity of attracting moths only in blue light with PBAN treatment. We consider this phenomenon is related to the physiology of the female moths' rut situation.</p> | |
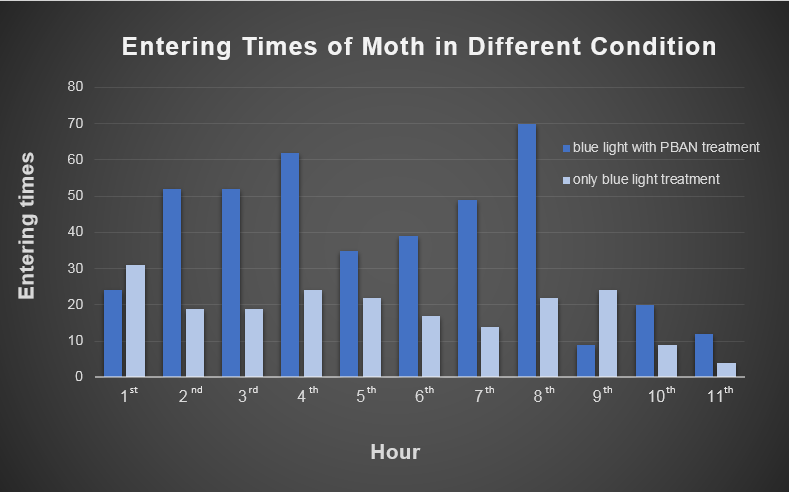
| - | + | [[File:Magic_Power_of_Our_Device.png|thumb|center|700px|Fig.1-1 Entering times is defined as the times that moths get into the device. The times of moths entering the device was recorded hourly for eleven hours, and can clearly see the magic power of our pyramidal device by using blue light with PBAN treatment.]] | |
| - | <p> | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <br> <br> | |
| - | <p> | + | <p>Our experiment can be divided into two categories. </p> |
| - | + | <p>1. PBAN Biobricks Tests: gene recombination and protein expression. </p> | |
| + | <p>2. Insect Tests: PBAN effect test, insect behavior test and device test. </p> | ||
| - | [[File:Average_GC_content.png|thumb|center|1200px| | + | </div></div> |
| + | <div class="li"><div class="card"> | ||
| + | |||
| + | ===PBAN Biobricks Tests=== | ||
| + | ====PBAN Gene Synthesis (Full Gene Sequence Design Process)==== | ||
| + | <p>To capture the harmful insects causing damage in agriculture, we first found 9 different kinds of PBAN peptide of harmful insects common in many places of the world from our long literature review. Next, we obtain the DNA sequences by reversely translating the peptide sequences of these PBANs from NCBI (ex: PBAN Spodoptera litura: '''http://www.ncbi.nlm.nih.gov/protein/AAK84160.1''' ) Finally, we '''modified every codon''' on the DNA sequence and '''designed the DNA sequence''' for ''E.coli'' to express a certain PBAN. </p> | ||
| + | <p>DNA Modification Process: </p> | ||
| + | <p>1. Avoid the rare codons of ''E.coli'', and choose high frequency codons. <br> | ||
| + | (Frequency Table Tool: '''http://www.genscript.com/cgi-bin/tools/codon_freq_table''')</p> | ||
| + | <p>Use [http://www.genscript.com/cgi-bin/tools/rare_codon_analysis Rare Codon Analysis Tool] to inspect if there is any problem to express our gene for ''E.coli.'' </p> | ||
| + | <p>2. Avoid choosing the same codon when modifying our designed gene sequence to prevent the ''E.coli'' from running out of nucleotides due to repeated use. </p> | ||
| + | <p>3. Avoid the start codon ATG in the middle of the coding sequence. </p> | ||
| + | |||
| + | |||
| + | <p>Take the PBAN of ''Spodoptera litura'' for example: </p> | ||
| + | [[File:PBAN_Spodoptera_litura_CAI_Value.png|thumb|center|1200px| Fig.2-1-1 The distribution of codon usage frequency along the length of your CDS to be expressed in your target host organism. Possibility of high protein expression level is correlated to the value of CAI - a CAI of 1.0 is considered to be ideal while a CAI of >0.8 is rated as good for expression in the desired expression organism. GenScript's OptimumGeneTM codon optimization tool can typically improve your sequence to reach a CAI of higher than 0.8 thus better chance of high level protein expression.]] | ||
| + | |||
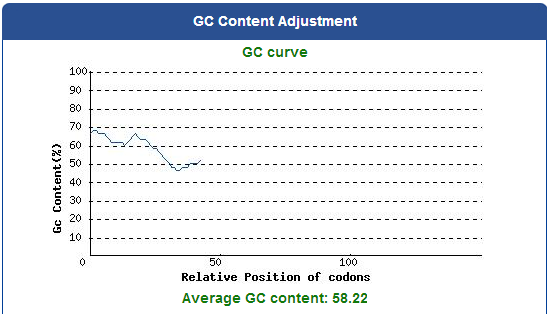
| + | [[File:Average_GC_content.png|thumb|center|1200px| Fig.2-1-2 The ideal percentage range of GC content is between 30% to 70%. Any peaks outside of this range will adversely affect transcription and translation efficiency.]] | ||
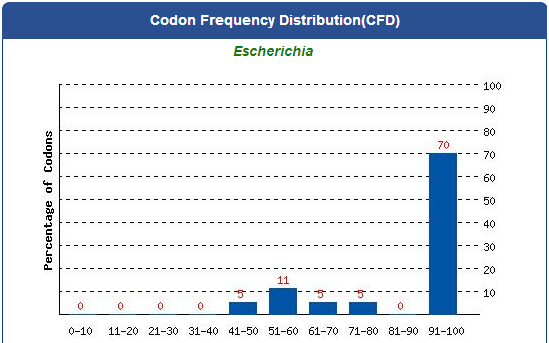
| - | [[File:Codon_Frequency_Distribution.png|thumb|center|1200px| | + | [[File:Codon_Frequency_Distribution.png|thumb|center|1200px| Fig.2-1-3 The percentage distribution of codons in computed codon quality groups. The value of 100 is set for the codon with the highest usage frequency for a given amino acid in the desired expression organism. Codons with values lower than 30 are likely to hamper the expression efficiency.]] |
<p>5. Add iGEM standard sequence in front of and at the back of our modified DNA sequence.</p> | <p>5. Add iGEM standard sequence in front of and at the back of our modified DNA sequence.</p> | ||
| - | <p>6. | + | <p>6. Synthesize the modified DNA sequence of PBANs in a gene synthesis company.</p> |
| - | ==== | + | ====PCR experiment of PBAN==== |
[[File:PBAN_Biobrick.png|link=|frameless|center|300px]] | [[File:PBAN_Biobrick.png|link=|frameless|center|300px]] | ||
| - | <p> | + | <p>After receiving the DNA sequences from the gene synthesis company, we recombined each PBAN gene to PSB1C3 backbones and conducted a PCR experiment to check the size of each of the PBANs. </p> |
| - | + | ||
| - | + | ||
| - | + | [[File:NEWWWWWW PBAN.png|center|thumb|650px|Fig.2-2-1 The PCR result of the 9 different kinds of PBAN. The DNA sequence length of PBANs are around 100-150 bp, so the PCR products should appear at 300-350 bp.<p> | |
| - | [[File: | + | Below are biobrick serial numbers of PBAN abbrevation:</p> |
| - | <p>In | + | BM: BBa_K1415001 MB: BBa_K1415002 AI: BBa_K1415003<p></p> |
| + | LD: BBa_K1415004 SL: BBa_K1415005 HAH: BBa_K1415006<p></p> | ||
| + | AS: BBa_K1415007 SI: BBa_K1415008 AA: BBa_K1415009<p></p> | ||
| + | ]] | ||
| + | <p>The DNA sequence length of the PBAN are around 100-150 bp. In this PCR experiment, the PBAN products size should be around 300-350 bp. Fig.2-2-1 showed the correct size of the PBAN, and proved that we '''successful ligated the PBAN DNA sequence onto an ideal backbone'''.</p> | ||
| - | [[File: | + | ====PBAN Peptide Check by SDS Protein Electrophoresis==== |
| + | [[File:Pcons+RBS+PBAN_Biobrick.png|link=|frameless|center|450px]] | ||
| + | <p>Moreover, to verify that all 9 kinds of PBAN can be expressed by the ''E.coli'', we conducted a '''SDS protein electrophoresis experiment'''. We first smashed the ''E.coli'' containing the PBAN with a sonicator and then took the supernatant divided from the bacterial pellet by centrifugation. Finally, we used the supernatant to run a SDS protein electrophoresis in a 20 % SDS gel.</p> | ||
| - | [[File: | + | [[File:NCTUSDS-1.png|thumb|center|700px|Fig.2-3-1 Protein Electrophoresis of P<sub>cons</sub> + RBS + 5 different kinds of PBAN (control: plasmid of P<sub>cons</sub>+RBS). Each peptide of PBAN is an around 30 amino acids, so we can see the band of PBANs at 2-4 kDa.<p> |
| + | Below are biobrick serial numbers of PBAN abbrevation:</p> | ||
| + | BM: BBa_K1415101 AA: BBa_K1415109 LD: BBa_K1415104 <p></p> | ||
| + | AS: BBa_K1415107 SL: BBa_K1415105 <p></p> | ||
| + | ]] | ||
| - | + | [[File:NCTUSDS-2.png|thumb|center|700px|Fig.2-3-2 Protein Electrophoresis of P<sub>cons</sub> + RBS + 4 different kinds of PBAN (control: plasmid of P<sub>cons</sub>+RBS). Each peptide of PBAN is an around 30 amino acids, so we can see the band of PBANs at 2-4 kDa.<p> | |
| + | Below are biobrick serial numbers of PBAN abbrevation:</p> | ||
| + | AI: BBa_K1415103 MB: BBa_K1415102 HAH: BBa_K1415106 <p></p>SI: BBa_K1415108<p></p> | ||
| - | ====Blue Light Fluorescence | + | ]] |
| + | |||
| + | These SDS PAGE results in Fig.2-3-1 showed that the '''bands are at 2-4 kDa''' for each of the PBANs, while the plasmid of P<sub>cons</sub>+RBS weren't there (the PBAN peptide is around 30 amino acids long). This result proves that the''' ''E.coli'' can produce the PBAN''' of our interest. | ||
| + | |||
| + | ====Blue Light Fluorescence Test==== | ||
[[File:Pcons+RBS+PBAN+RBS+BFP+Ter_Biobrick.png|link=|frameless|center|650px]] | [[File:Pcons+RBS+PBAN+RBS+BFP+Ter_Biobrick.png|link=|frameless|center|650px]] | ||
| - | <p> | + | <p>To predict the PBAN expression in ''E.coli'' by computer modeling, we next tested PBAN BFP biobricks. We obtained the average expressive value of the blue fluorescence in the biobrick part (above) and also the control part of P<sub>cons</sub> + RBS + BFP + Ter. Therefore, we can use the average value to generate predictions of the PBAN expression in ''E.coli''. |
| - | [https://2014.igem.org/Team:NCTU_Formosa/modeling#Modeling_for_biobricks ( '' | + | [https://2014.igem.org/Team:NCTU_Formosa/modeling#Modeling_for_biobricks (''See more details in our Modeling Page'').] |
| - | + | Below is the '''blue fluorescence expression''' curve in a long period of time. We used these data to predict the PBAN expression in ''E.coli''. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | === | + | [[File:PBAN_Fluorescence_Value.jpg|thumb|center|900px|Fig.2-4-1 The blue light fluorescence expression curve of ''E.coli'' containing P<sub>cons</sub> + RBS + 9 different kinds of PBAN + RBS + BFP + Ter plasmid (control: competent cells that cannot emit blue light). Below are biobrick serial numbers of the PBAN abbreviations:</p> |
| - | [[File:Autoclave.png|thumb|center| | + | SL: BBa_K1415205 BM: BBa_K1415201 MB: BBa_K1415202<p></p> |
| - | + | AI: BBa_K1415203 LD: BBa_K1415204 HAH:BBa_K1415206<p></p> | |
| + | AS: BBa_K1415207 SI: BBa_K1415208 AA: BBa_K1415209<p></p> ]] | ||
| + | In Fig.2-4-2, we can clearly see that the blue fluorescence expressed by the ''E.coli'' is different from the control without BFP expressed. Also, we find that different kinds of PBAN has different expression in ''E.coli''. We think because different PBAN has '''different codons''' on DNA sequence, '''different PBAN peptide''' or PBAN plasmid will be expressed or replicated in different amount. | ||
| + | [[File:Blue_light_flourescence_of_9_different_kinds_of_PBAN.png|thumb|center|800px| Fig.2-4-2 Blue Fluorescence of P<sub>cons</sub> + RBS + 9 different kinds of PBAN + RBS + BFP + Ter plasmid (control: ''E.coli'' containg P<sub>cons</sub>+RBS Plasmid). Below are biobrick serial numbers of the PBAN abbreviations:<br> | ||
| + | <p style="text-align:left;">SL: BBa_K1415205 BM: BBa_K1415201 MB: BBa_K1415202</p> | ||
| + | <p style="text-align:left;">AI: BBa_K1415203 LD: BBa_K1415204 HAH:BBa_K1415206</p> | ||
| + | <p style="text-align:left;">AS: BBa_K1415207 SI: BBa_K1415208 AA: BBa_K1415209</p> ]] | ||
| + | </div></div> | ||
| + | <div class="li"><div class="card"> | ||
| + | |||
| + | ===Obtaining PBAN from ''E.coli''=== | ||
| + | [[File:Autoclave.png|thumb|center|700px|Fig.3-1-1 The Process of Obtaining PBAN from ''E.coli''. We first culture the ''E.coli'' that contains our constructed plasmid of P<sub>cons</sub> + RBS + One Kind of PBAN for 12 hr. Second, We smash ''E.coli'' with a sonicator to divide the PBAN from the cell debris pellets. Finally, we autoclave the supernatant to avoid biosafet issues.]] | ||
| + | <p>In order to obtain PBAN from our ''E.coli'', we first culture the ''E.coli'' that contains our constructed plasmid of P<sub>cons</sub> + RBS + One Kind of PBAN in best time condition at which [https://2014.igem.org/File:PBAN_OD600_Value.jpg the blue light fluorescence expression] reaches the maximum value. Then, we centrifuged the bacterial solution to pour-off the LB broth and resuspend the ''E.coli'' pellet with pure water later for the next step. From this step, we can obtain a LB-free solution, which is better for our insect experiment. Then, we '''smash''' the ''E.coli'' with a sonicator and '''dilute''' this PBAN solution with 250 ml pure water in a serum bottle. Finally, we '''sterlized''' the PBAN solution to avoid biosafety issues. As we know, PBAN is a very '''simple and short''' peptide so it will not be degraded after the autoclave treatment. Since only a very small amount (10 pmol) of PBAN will be enough to stimulate the maximum production of pheromone, therefore, we don't have to worry that our PBAN concentration will be inadequate after diluting with 250 ml pure water. Overall, we can simply use the sterilized solution without purification, following recommended culturing conditions. </p> | ||
| + | |||
| + | |||
| + | |||
| + | [[File:Cultivation_of_E.coli..png|290px|thumb|left||Fig.3-1-2<br> We culture the ''E.coli'' in best time condition to have the maximum expression of PBAN.<br> | ||
| + | Below are biobrick serial numbers of PBAN abbrevation:<br> | ||
| + | <span style="text-align:left;">MB: BBa_K1415102<br>HAH: BBa_K1415106<br>SL: BBa_K1415105</span>]] | ||
| + | |||
| + | [[File:Smash_E.coli._with_Sonicator.png|290px|thumb|right||Fig.3-1-4<br> We smash the ''E.coli'' with a sonicator.<br> | ||
| + | Below are biobrick serial numbers of PBAN abbrevation:<br> | ||
| + | <span style="text-align:left;">MB: BBa_K1415102<br>HAH: BBa_K1415106<br>SL: BBa_K1415105</span>]] | ||
| + | |||
| + | [[File:Centrifuge_&_Pour.png|290px|thumb|center||Fig.3-1-3<br> We centrifuged the bacterial solution to pour LB broth.<br> | ||
| + | Below are biobrick serial numbers of PBAN abbrevation:<br> | ||
| + | <span style="text-align:left;">MB: BBa_K1415102<br>HAH: BBa_K1415106<br>SL: BBa_K1415105</span>]] | ||
| + | |||
| + | [[File:PBAN_Dilution.png|290px|thumb|left||Fig.3-1-5<br> We dilute this PBAN solution with 250ml pure water in serum bottle.<br> | ||
| + | Below are biobrick serial numbers of PBAN abbrevation:<br> | ||
| + | <span style="text-align:center;">MB: BBa_K1415102<br>HAH: BBa_K1415106<br>SL: BBa_K1415105</span>]] | ||
| + | [[File:Autoclave.JPG|290px|thumb|right||Fig.3-1-6<br> We sterlized PBAN solution to avoid biosafety issues.]] | ||
| + | |||
| + | |||
| + | </div></div> | ||
| + | <div class="li"><div class="card"> | ||
===Insect Tests=== | ===Insect Tests=== | ||
====Behavior of Target Insects After PBAN Treatment==== | ====Behavior of Target Insects After PBAN Treatment==== | ||
<p> | <p> | ||
| - | To | + | To investigate what behavior the female moth would show after sucking PBAN, we put one female moth into a beaker for observation. The beaker is divided into two parts by plastic wrap. The bottom part contains the PBAN solution we prepared, and the upper part is the space for the moth to stay(showed in Fig.4-2-3). We soaked cotton that spans the entire length of the beaker with the PBAN solution and sprinkle it with sugar. This way, the moth can suck on the PBAN without drowning in PBAN solution. After all the equipment is set, we put the female moth into the upper part of the beaker. At the time, we started filming as soon as we observed the female moth showing obvious behaviors of '''sexual stimulation''' such as flapping their wings.</p> |
| + | <p>The activeness of the 9 kinds of target insects changes with the '''seasons'''. Our observation was made around September to October, which is the most active time for ''Spodoptera litura, Mamestra brassicae'' and ''Helicoverpa armigera Hubner''. Therefore, in this observation, we caught these three kinds of moth in Sunnymorning organic farm as sample moths. </p> | ||
<p> | <p> | ||
| - | We | + | We observed that the moth could absorb the PBAN in the solution through suck, and that the PBAN could '''stimulate''' the moth's '''pheromone gland''' to produce pheromone.</p> <p>'''As soon as the moth is sexually excited, it would flap its wings rapidly and move its tail slightly upward.'''</p> |
| - | </p> | + | |
<div> | <div> | ||
{{:Team:NCTU Formosa/source/result-video}} | {{:Team:NCTU Formosa/source/result-video}} | ||
</div> | </div> | ||
| - | These | + | These vidoes show the behaviors of 3 different kinds of female moths after sucking their separate PBANs. Each of the moths clearly became '''excited and all flapped their wings rapidly'''. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ====Effective Attraction after PBAN Treatment==== | |
| - | + | After observing how PBAN treatment affects the behavior of female moths, we want to check the '''attractive effect''' of the moths. We expected that, after sucking the PBAN, the female moth would not only become excited, '''flap its wings''', but also actually attract male moths to '''aggregate''' together. We used two beakers which are the same as what we used in the former experiment. One contained PBAN solution and the other contained only sucrose solution as control. We first put one beaker at one side and the other at the opposite side in the moth box (shown in Fig.4-2-1). Then we put two female moths in each beaker and at least 100 male moths in the moth box. This time, we did a long time observation and took a picture with our camera. In Fig.4-2-1, the female moth sucked the PBAN then attracted more male moths than the one sucking the sucrose solution. Thus, Fig.4-2-1 can prove that the female moth sucked our PBAN then released a large amount of sex pheromone to attract many male moths. In addition, we conducted a simple test to compare the luring effect of female moths sucking PBAN solution with that of sucrose solution (moth's favorite food). Again, we can see the conspicuous effect. | |
| + | [[File:PBAN_Basic_Effect.jpg|thumb|left|200px|Fig.4-2-1 Negative Control: Female moths suck sucrose solution without PBAN (number = 0). Experiment: PBAN sucking of Female moths (number = 11). In this picture, we can see that the female moths sucking PBAN solution can release much sex pheromone, and attract many male moths. ]] | ||
| + | [[File:Before_&_After.jpg|thumb|center|700px|Fig.4-2-2 Negative Control: sucrose solution, Experiment: Female moth eating PBAN solution. Also, we can see the PBAN effect again from this picture.]] | ||
| + | [[File:2014_NCTU_Formosa_beaker_layer.jpg|thumb|center|700px|Fig.4-2-3 The container we used to contain female moths with PBAN solution, we designed it with beaker.]] | ||
| - | + | ====''Spodoptera Litura'''s Preference for Temperature and Light ==== | |
| - | <p>Temperature is | + | As we all know, light can attract many kinds of harmful insects. |
| - | [[File:Moth's_Behavior,_Temperature_and_Light.png|thumb|center|800px| | + | <p>Temperature is another environmental factor which the farmer can not change practically. We want to use computer '''modeling''' to explore in depth the moths' preferences for different combinations of '''light and temperature''' conditions. In the future, we hope that farmers can choose the appropriate light according to temperature condition when using our device. For this purpose, we chose the average temperature range in Taiwan in a year, and most common harmful insect, ''Spodoptera Litura'' to conduct this test (Fig.4-3-1 below), which we used to model the moths' preferences for different combinations of light and temperature conditions with ANFIS (see detail in the [https://2014.igem.org/Team:NCTU_Formosa/modeling#Device_modeling modeling of device] page). </p> |
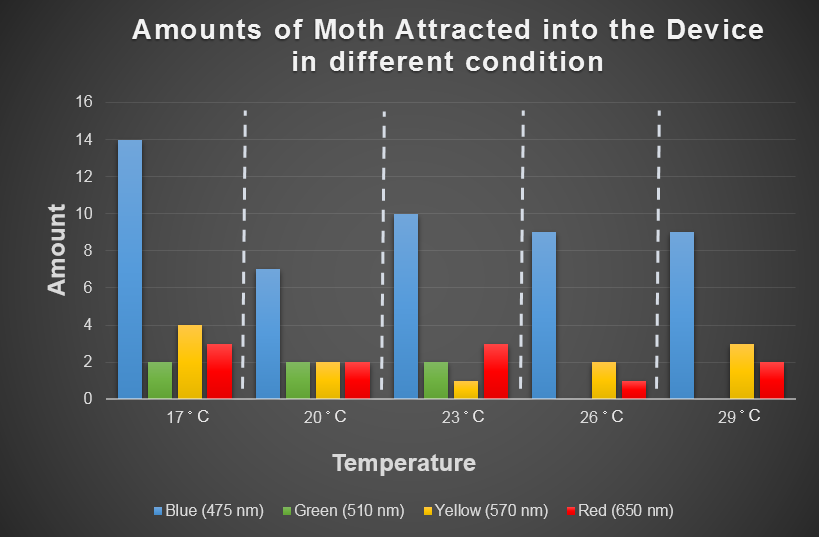
| + | [[File:Moth's_Behavior,_Temperature_and_Light.png|thumb|center|800px|Fig.4-3-1 Result of 30 repeats of ''Spodoptera Litura'''s preference testing for temperature and light (there are some moths staying in the bottom in every test). In this table, we can see blue light has a consistent attractive effect ''Spodoptera Litura'' under different temperature condition. ]] | ||
| + | Fig.4-3-1 shows blue light has consistent attraction to our target harmful moth, ''Spodoptera Litura'', in any temperature condition. Thus, we decided to use blue LED light in our device design. | ||
| + | [[File:CCW_Model.JPG|thumb|center|600px|Fig.4-3-2 Modeling tool (CCW No.1) for this test. We use blue, green, red, yellow, and positive control: white at the same time under different temperature conditions for our device modeling testing. ''Spodoptera Litura''s were packed into the central barrel. Every test was followed by continuous beating on the barrel for 5 min in order to make the moths fly. ]] | ||
| - | |||
| - | |||
<div></div> | <div></div> | ||
Latest revision as of 03:58, 18 October 2014
Contents |
Magic Power of Our Pyramidal Device
We develop a powerful and specific insect attractive device which combine the use of blue light and PBAN. The video shows that our device successfully and effectively attract more insects. In this video:
1. We used our pyramidal device, biobrick part [http://parts.igem.org/Part:BBa_K1415105 BBa_K1415105: Pcons+RBS+PBAN(Spodoptera litura)] and its target insect Spodoptera litura inside an arcrylic chamber for this expiriment.
2. The female moth is fed with PBAN in a small beaker covering with plastic wrap. Soon, the female flaps its wings frantically, a sign of sexual stimulation. From this point on, it starts to release pheromones.
3. We transferred the beaker into our device then placed the device in an acrylic chamber(moth box) with approximately 150 male moths inside to conduct the test. We kept the chamber dark to ensure that blue light would be the only light source inside. We recorded the number of insects entering the device per hour for eleven hours.
Fig.1-1 shows the entering times of moths in different conditions. Entering times is defined as the times that moths get into the device. In this experiment, the moth might get out of our device, since we were only using a simple model device without trap, such as nets for keeping the moths. However, we can still clearly see the magic power of our pyramidal device by using blue light with PBAN treatment, which attracted more moths to get into the device than the other experiment (only blue light treatment). In addition, we can even observe a unique periodicity of attracting moths only in blue light with PBAN treatment. We consider this phenomenon is related to the physiology of the female moths' rut situation.
Our experiment can be divided into two categories.
1. PBAN Biobricks Tests: gene recombination and protein expression.
2. Insect Tests: PBAN effect test, insect behavior test and device test.
PBAN Biobricks Tests
PBAN Gene Synthesis (Full Gene Sequence Design Process)
To capture the harmful insects causing damage in agriculture, we first found 9 different kinds of PBAN peptide of harmful insects common in many places of the world from our long literature review. Next, we obtain the DNA sequences by reversely translating the peptide sequences of these PBANs from NCBI (ex: PBAN Spodoptera litura: http://www.ncbi.nlm.nih.gov/protein/AAK84160.1 ) Finally, we modified every codon on the DNA sequence and designed the DNA sequence for E.coli to express a certain PBAN.
DNA Modification Process:
1. Avoid the rare codons of E.coli, and choose high frequency codons.
(Frequency Table Tool: http://www.genscript.com/cgi-bin/tools/codon_freq_table)
Use [http://www.genscript.com/cgi-bin/tools/rare_codon_analysis Rare Codon Analysis Tool] to inspect if there is any problem to express our gene for E.coli.
2. Avoid choosing the same codon when modifying our designed gene sequence to prevent the E.coli from running out of nucleotides due to repeated use.
3. Avoid the start codon ATG in the middle of the coding sequence.
Take the PBAN of Spodoptera litura for example:
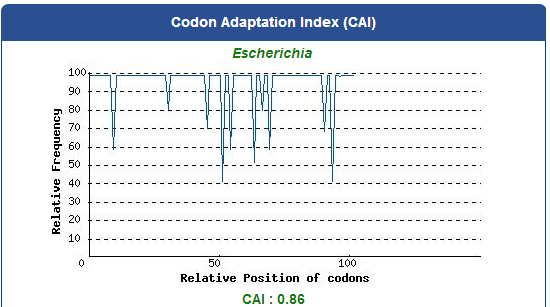
5. Add iGEM standard sequence in front of and at the back of our modified DNA sequence.
6. Synthesize the modified DNA sequence of PBANs in a gene synthesis company.
PCR experiment of PBAN

After receiving the DNA sequences from the gene synthesis company, we recombined each PBAN gene to PSB1C3 backbones and conducted a PCR experiment to check the size of each of the PBANs.
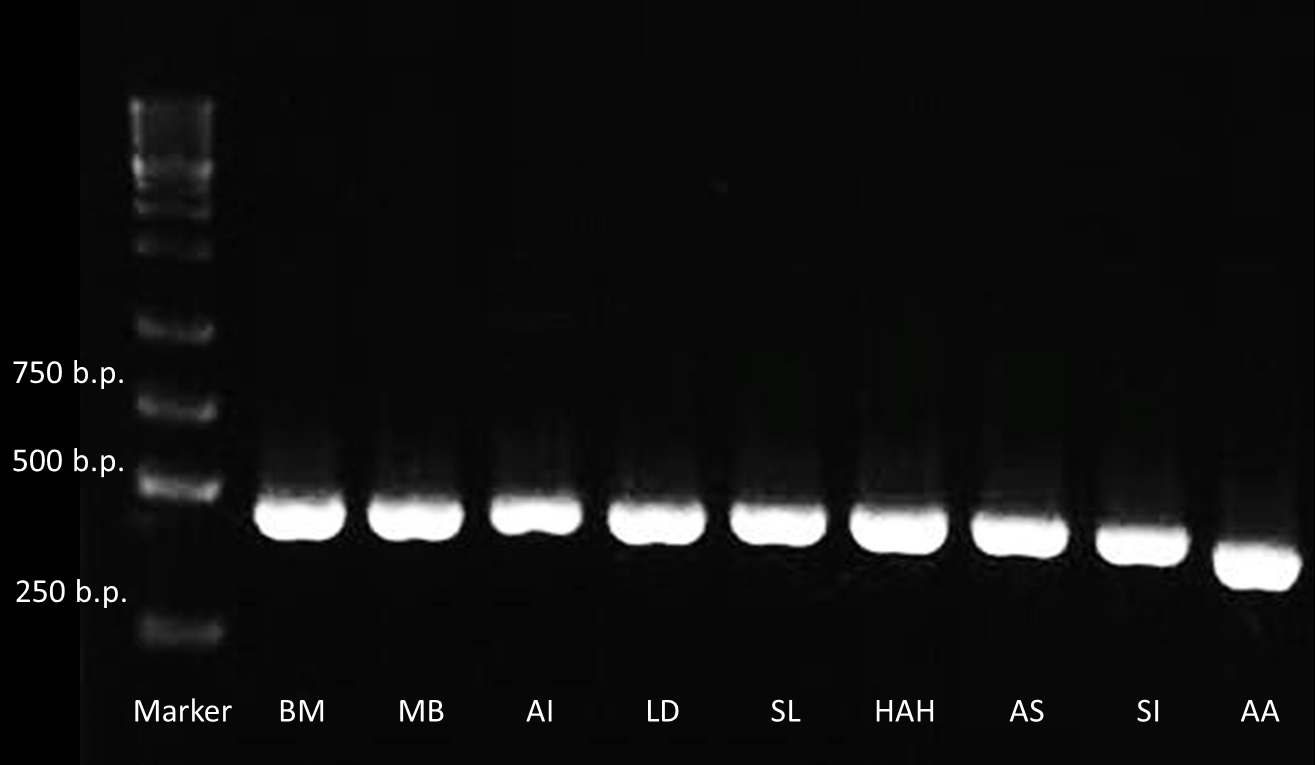
Below are biobrick serial numbers of PBAN abbrevation:
BM: BBa_K1415001 MB: BBa_K1415002 AI: BBa_K1415003 LD: BBa_K1415004 SL: BBa_K1415005 HAH: BBa_K1415006 AS: BBa_K1415007 SI: BBa_K1415008 AA: BBa_K1415009The DNA sequence length of the PBAN are around 100-150 bp. In this PCR experiment, the PBAN products size should be around 300-350 bp. Fig.2-2-1 showed the correct size of the PBAN, and proved that we successful ligated the PBAN DNA sequence onto an ideal backbone.
PBAN Peptide Check by SDS Protein Electrophoresis

Moreover, to verify that all 9 kinds of PBAN can be expressed by the E.coli, we conducted a SDS protein electrophoresis experiment. We first smashed the E.coli containing the PBAN with a sonicator and then took the supernatant divided from the bacterial pellet by centrifugation. Finally, we used the supernatant to run a SDS protein electrophoresis in a 20 % SDS gel.

Below are biobrick serial numbers of PBAN abbrevation:
BM: BBa_K1415101 AA: BBa_K1415109 LD: BBa_K1415104 AS: BBa_K1415107 SL: BBa_K1415105
Below are biobrick serial numbers of PBAN abbrevation:
AI: BBa_K1415103 MB: BBa_K1415102 HAH: BBa_K1415106 SI: BBa_K1415108These SDS PAGE results in Fig.2-3-1 showed that the bands are at 2-4 kDa for each of the PBANs, while the plasmid of Pcons+RBS weren't there (the PBAN peptide is around 30 amino acids long). This result proves that the E.coli can produce the PBAN of our interest.
Blue Light Fluorescence Test

To predict the PBAN expression in E.coli by computer modeling, we next tested PBAN BFP biobricks. We obtained the average expressive value of the blue fluorescence in the biobrick part (above) and also the control part of Pcons + RBS + BFP + Ter. Therefore, we can use the average value to generate predictions of the PBAN expression in E.coli. (See more details in our Modeling Page). Below is the blue fluorescence expression curve in a long period of time. We used these data to predict the PBAN expression in E.coli.
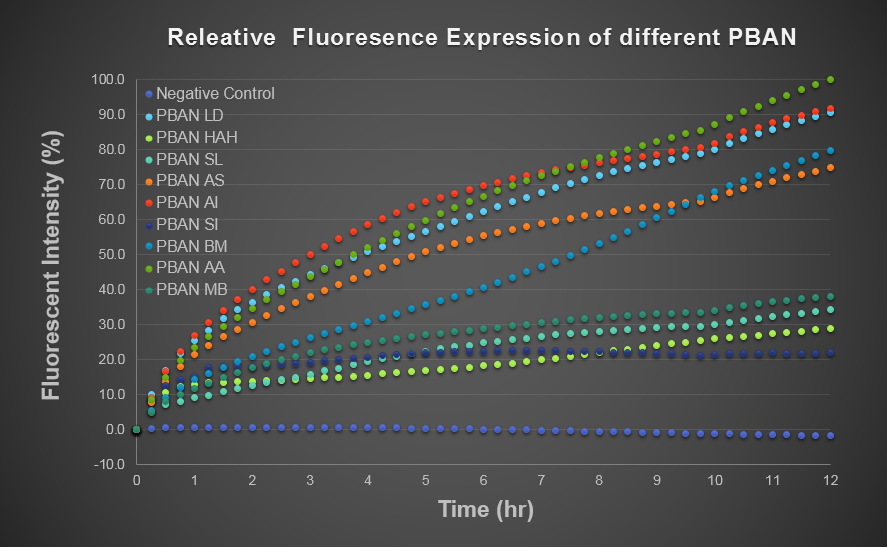
In Fig.2-4-2, we can clearly see that the blue fluorescence expressed by the E.coli is different from the control without BFP expressed. Also, we find that different kinds of PBAN has different expression in E.coli. We think because different PBAN has different codons on DNA sequence, different PBAN peptide or PBAN plasmid will be expressed or replicated in different amount.

SL: BBa_K1415205 BM: BBa_K1415201 MB: BBa_K1415202
AI: BBa_K1415203 LD: BBa_K1415204 HAH:BBa_K1415206
AS: BBa_K1415207 SI: BBa_K1415208 AA: BBa_K1415209
Obtaining PBAN from E.coli

In order to obtain PBAN from our E.coli, we first culture the E.coli that contains our constructed plasmid of Pcons + RBS + One Kind of PBAN in best time condition at which the blue light fluorescence expression reaches the maximum value. Then, we centrifuged the bacterial solution to pour-off the LB broth and resuspend the E.coli pellet with pure water later for the next step. From this step, we can obtain a LB-free solution, which is better for our insect experiment. Then, we smash the E.coli with a sonicator and dilute this PBAN solution with 250 ml pure water in a serum bottle. Finally, we sterlized the PBAN solution to avoid biosafety issues. As we know, PBAN is a very simple and short peptide so it will not be degraded after the autoclave treatment. Since only a very small amount (10 pmol) of PBAN will be enough to stimulate the maximum production of pheromone, therefore, we don't have to worry that our PBAN concentration will be inadequate after diluting with 250 ml pure water. Overall, we can simply use the sterilized solution without purification, following recommended culturing conditions.
Insect Tests
Behavior of Target Insects After PBAN Treatment
To investigate what behavior the female moth would show after sucking PBAN, we put one female moth into a beaker for observation. The beaker is divided into two parts by plastic wrap. The bottom part contains the PBAN solution we prepared, and the upper part is the space for the moth to stay(showed in Fig.4-2-3). We soaked cotton that spans the entire length of the beaker with the PBAN solution and sprinkle it with sugar. This way, the moth can suck on the PBAN without drowning in PBAN solution. After all the equipment is set, we put the female moth into the upper part of the beaker. At the time, we started filming as soon as we observed the female moth showing obvious behaviors of sexual stimulation such as flapping their wings.
The activeness of the 9 kinds of target insects changes with the seasons. Our observation was made around September to October, which is the most active time for Spodoptera litura, Mamestra brassicae and Helicoverpa armigera Hubner. Therefore, in this observation, we caught these three kinds of moth in Sunnymorning organic farm as sample moths.
We observed that the moth could absorb the PBAN in the solution through suck, and that the PBAN could stimulate the moth's pheromone gland to produce pheromone.
As soon as the moth is sexually excited, it would flap its wings rapidly and move its tail slightly upward.
These vidoes show the behaviors of 3 different kinds of female moths after sucking their separate PBANs. Each of the moths clearly became excited and all flapped their wings rapidly.
Effective Attraction after PBAN Treatment
After observing how PBAN treatment affects the behavior of female moths, we want to check the attractive effect of the moths. We expected that, after sucking the PBAN, the female moth would not only become excited, flap its wings, but also actually attract male moths to aggregate together. We used two beakers which are the same as what we used in the former experiment. One contained PBAN solution and the other contained only sucrose solution as control. We first put one beaker at one side and the other at the opposite side in the moth box (shown in Fig.4-2-1). Then we put two female moths in each beaker and at least 100 male moths in the moth box. This time, we did a long time observation and took a picture with our camera. In Fig.4-2-1, the female moth sucked the PBAN then attracted more male moths than the one sucking the sucrose solution. Thus, Fig.4-2-1 can prove that the female moth sucked our PBAN then released a large amount of sex pheromone to attract many male moths. In addition, we conducted a simple test to compare the luring effect of female moths sucking PBAN solution with that of sucrose solution (moth's favorite food). Again, we can see the conspicuous effect.
Spodoptera Litura's Preference for Temperature and Light
As we all know, light can attract many kinds of harmful insects.
Temperature is another environmental factor which the farmer can not change practically. We want to use computer modeling to explore in depth the moths' preferences for different combinations of light and temperature conditions. In the future, we hope that farmers can choose the appropriate light according to temperature condition when using our device. For this purpose, we chose the average temperature range in Taiwan in a year, and most common harmful insect, Spodoptera Litura to conduct this test (Fig.4-3-1 below), which we used to model the moths' preferences for different combinations of light and temperature conditions with ANFIS (see detail in the modeling of device page).
Fig.4-3-1 shows blue light has consistent attraction to our target harmful moth, Spodoptera Litura, in any temperature condition. Thus, we decided to use blue LED light in our device design.

 "
"