Team:Paris Saclay/Project/Salicylate Inducible System
From 2014.igem.org
MarieThanh (Talk | contribs) (→Salicylate Inducible System) |
(→Alternative pathway) |
||
| (178 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:Paris_Saclay/project_header}} | {{Team:Paris_Saclay/project_header}} | ||
| - | = | + | =Lemon Appearance And Ripening= |
| - | == | + | ==Introduction== |
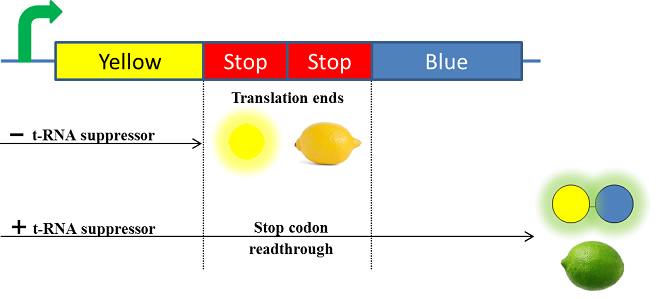
| - | + | As everyone knows, lemon is a fruit that can display a yellow color when it is ripe and a green color when it is not. Firstly, we chose to use chromoproteins to express these colors in E. coli. Chromoproteins are reflective proteins that contain a pigmented prosthetic group (for example, iron for the haemoglobin) and do not need to be excited to be seen. Although several chromoproteins have been synthesized in the past years, there is no known green chromoprotein yet synthesized in iGEM. We aim to resolve this by fusing a yellow chromoprotein with a blue one that hopefully will display a green color. This construction will be referred as the green fusion chromoprotein. | |
| - | + | Secondly, in order to make our lemon ripe like a real lemon, we decided to take advantage of the designing of the fusion protein by using a translational suppression system. Indeed, we plan to add amber codons within the linker separating the yellow and the blue chromoproteins. Therefore, the expression of a suppressor t-RNA will suppress amber codons allowing the translation of the green fusion chromoprotein. Conversely, the down regulation of the suppressor t-RNA through time will allow bacteria switch from green to yellow, thus simulating the ripening of a real lemon. This system will be referred to as the color switch system. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | [[File:Paris_Saclay_project-lemon_appearance_&_rippening.jpg|center]] | |
| - | + | == The green fusion chromoprotein design == | |
| - | + | ||
| + | ====Promoter & RBS==== | ||
| + | |||
| + | Because the ripening of the lemon will be achieved by the regulation of the expression of the suppressor t-RNA, the fusion chromoprotein has been placed under control of a constitutive promoter. To ensure that E. coli produces a robust color, we chose the RBS BBa_B0034 (strength = 1) and the “consensus” promoter BBa_J23119. | ||
| + | |||
| + | ====Chromoproteins==== | ||
| + | |||
| + | Uppsala iGEM team 2013 already designed several chromoproteins such as the FwYellow (BBa_K1033910) and the AeBlue chromoproteins (BBa_K1033902) [1]. We chose to fuse these chromoproteins so it should produce a green color. | ||
| + | |||
| + | ====Linker==== | ||
| + | |||
| + | Linkers are short sequences usually containing flexible amino-acid such as glycine and serine to ensure that two proteins domain are correctly folded and avoid steric issues. Thus, a linker of 12 amino acids that repeat a motif of glycine-glycine-serine-glycine (BBa_K243006) has been used to separate both FwYellow and AeBlue chromoproteins. Furthermore, to permit the color switch by the translational suppression system, we plan to replace two serine codons from the linker by amber codons (TAG). Choosing to add two amber codons would ensure a low stop codon readthrough in absence of the t-RNA. | ||
| + | |||
| + | ====Terminator==== | ||
| + | |||
| + | To end the transcription of the fusion chromoprotein, a commonly used terminator BBa_B0010 is added. | ||
| + | |||
| + | ==The Color Switch system design== | ||
| + | |||
| + | ====The supD suppressor t-RNA==== | ||
| - | |||
| - | |||
Given that the linker that separates both FwYellow and AeBlue chromoproteins is composed of serine and glycine amino-acid, we needed a t-RNA suppressor that could encode one of these amino-acids to not alter the properties of the linker. | Given that the linker that separates both FwYellow and AeBlue chromoproteins is composed of serine and glycine amino-acid, we needed a t-RNA suppressor that could encode one of these amino-acids to not alter the properties of the linker. | ||
As we were looking for a t-RNA suppressor, we found that the iGEM Beijing 2009 team already worked on a translational suppression system. They worked with the supD suppressor t-RNA that encodes a serine making it an ideal candidate for our project. They placed supD under control of a salicylate inducible promoter Psal to suppress amber codon they had introduced in the T7 polymerase sequence. In turn, the T7 polymerase can express the GFP output gene. Their results show that supD does not induce bacteria lethality so such a system could be used [2]. | As we were looking for a t-RNA suppressor, we found that the iGEM Beijing 2009 team already worked on a translational suppression system. They worked with the supD suppressor t-RNA that encodes a serine making it an ideal candidate for our project. They placed supD under control of a salicylate inducible promoter Psal to suppress amber codon they had introduced in the T7 polymerase sequence. In turn, the T7 polymerase can express the GFP output gene. Their results show that supD does not induce bacteria lethality so such a system could be used [2]. | ||
| - | ===Transcription factor sensible to salicylate=== | + | |
| + | ====Transcription factor sensible to salicylate==== | ||
| + | |||
The nahR gene is involved in the degradation of the naphthalene pollutant in Pseudomonas putida. This gene encodes a transcriptional regulator that is induce by salicylate and thus bind nah or sal promoters. The BBa_K228004 Biobrick® contains the nahR gene under control of a constitutive promoter and the salicylate promoter (Psal). Thus, we plan to place supD under control of the Psal promoter. | The nahR gene is involved in the degradation of the naphthalene pollutant in Pseudomonas putida. This gene encodes a transcriptional regulator that is induce by salicylate and thus bind nah or sal promoters. The BBa_K228004 Biobrick® contains the nahR gene under control of a constitutive promoter and the salicylate promoter (Psal). Thus, we plan to place supD under control of the Psal promoter. | ||
| - | ===Color switch mechanism=== | + | |
| - | + | ====Color switch mechanism==== | |
| + | |||
| + | In the beginning, salicylate concentration will be maximal into the agar media so that supD will be expressed and so the green fusion chromoprotein: bacteria will display a green color. However, as bacteria grow into agar, less salicylate will remain available into the media. Thus, the decrease of the nahR-salicylate complex amount within bacteria will lead to supD downregulation through time. In turn, decrease of supD amount will result in less codon readthrough and so less translation of the green fusion protein and more translation of the yellow chromoprotein. As a result, bacteria will gradually change from green to yellow. | ||
| + | |||
| + | [[File:Paris_Saclay_project-ripening-debut.jpg.png|700px|center|At the beginning|link=https://static.igem.org/mediawiki/2014/e/ea/Paris_Saclay_project-ripening-debut.jpg.png]] | ||
| + | |||
| + | |||
| + | <hr> | ||
| + | |||
| + | |||
| + | [[File:Paris_Saclay_project-ripening-milieu.png|center|700px|Ripening process|link=https://static.igem.org/mediawiki/2014/2/26/Paris_Saclay_project-ripening-milieu.png]] | ||
| + | |||
| + | |||
| + | <hr> | ||
| + | |||
| + | |||
| + | [[File:Paris_Saclay_project-ripening-fin.png|center|700px|At the end|link=https://static.igem.org/mediawiki/2014/4/48/Paris_Saclay_project-ripening-fin.png]] | ||
| + | |||
| + | ==Results== | ||
| + | To save time the fusion chromoprotein gene was ordered to Lifetechnologies® company. It includes the suffix, the promoter, the RBS, the FwYellow’s open reading frame, the linker, the AeBlue’s open reading frame, the terminator and the prefix. As a first step, the linker did not bear the two amber codons so that we could know if the fusion chromoprotein is functional or not. | ||
| + | |||
| + | ====Cloning in pGEMTeasy==== | ||
| + | |||
| + | After receiving it, the fusion chromoprotein was amplified by PCR and then cloned into pGEMTeasy. Unfortunately, after transforming E.coli and plating the tranformation mix, no colonies display any color. We sequenced the gene cloned to ensure the absence of mutation. The simplest explanation is that the fusion protein is not anymore functional. One may suggest that the fact the chromoproteins need to be in a multimeric form to be functional, prevent the fusion protein to be functional. | ||
| + | |||
| + | As the fusion chromoprotein did not work, the colour switch system that we planned to do is no longer valid. | ||
| + | |||
| + | ==Alternative pathway== | ||
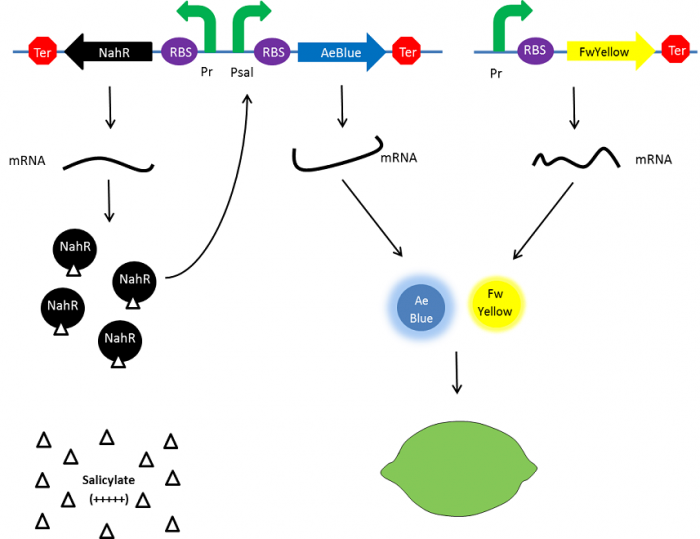
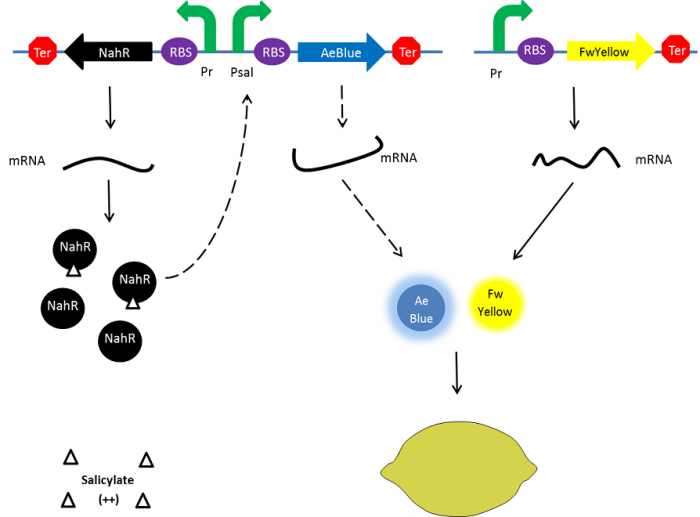
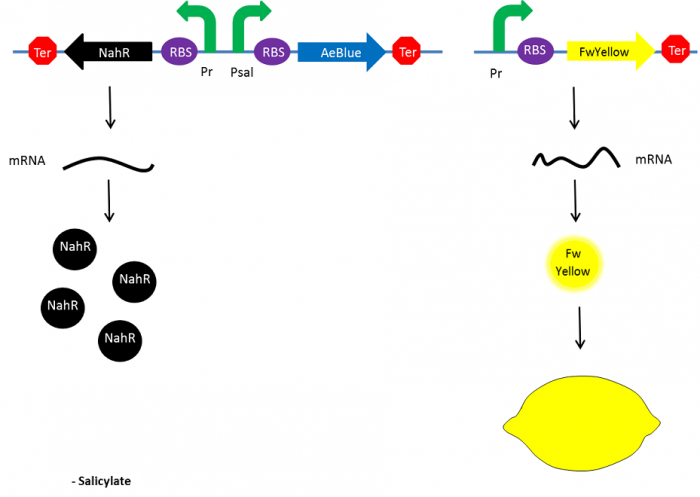
| + | To resolve this issue, we planned to separately express both FwYellow and AeBlue in bacteria. We decided to place the FwYellow under control of a constitutive promoter whereas AeBlue will be place under the Psal inducible promoter, as mentioned above. | ||
| + | As previously, the beginning, salicylate concentration is maximal into the agar media leading so both chromoprotein will be expressed. Assuming that the presence of both chromoprotein in the cytoplasm will display a green color, bacteria will be green. However, as bacteria grow into the agar media, the amount of nahR-salicylate complex will decrease through time. Thus, the expression of the AeBlue will decrease whereas the expression of FwYellow will remain equal. As a result, bacteria will gradually change from green to yellow. | ||
| + | |||
| + | |||
| + | [[File:Paris_Saclay_project-ripening-alternative-debut.png|700px|center|At the beginning|link=https://static.igem.org/mediawiki/2014/f/f9/Paris_Saclay_project-ripening-alternative-debut.png]] | ||
| + | |||
| + | |||
| + | <hr> | ||
| + | |||
| + | |||
| + | [[File:Paris_Saclay_project-ripening-alternative-milieu.png|700px|center|Ripening process|link=https://static.igem.org/mediawiki/2014/8/88/Paris_Saclay_project-ripening-alternative-milieu.png]] | ||
| + | |||
| + | |||
| + | <hr> | ||
| + | |||
| + | |||
| + | [[File:Paris_Saclay_project-ripening-alternative-fin.png|700px|center|At the end|link=https://static.igem.org/mediawiki/2014/7/75/Paris_Saclay_project-ripening-alternative-fin.png]] | ||
| + | |||
| + | ==References== | ||
| + | |||
| + | [1] https://2013.igem.org/Team:Uppsala/chromoproteins | ||
| + | |||
| + | [2] https://2009.igem.org/Team:PKU_Beijing/Parts_Characterization/BBa_K228258 | ||
{{Team:Paris_Saclay/default_footer}} | {{Team:Paris_Saclay/default_footer}} | ||
Latest revision as of 03:26, 18 October 2014

Contents |
Lemon Appearance And Ripening
Introduction
As everyone knows, lemon is a fruit that can display a yellow color when it is ripe and a green color when it is not. Firstly, we chose to use chromoproteins to express these colors in E. coli. Chromoproteins are reflective proteins that contain a pigmented prosthetic group (for example, iron for the haemoglobin) and do not need to be excited to be seen. Although several chromoproteins have been synthesized in the past years, there is no known green chromoprotein yet synthesized in iGEM. We aim to resolve this by fusing a yellow chromoprotein with a blue one that hopefully will display a green color. This construction will be referred as the green fusion chromoprotein.
Secondly, in order to make our lemon ripe like a real lemon, we decided to take advantage of the designing of the fusion protein by using a translational suppression system. Indeed, we plan to add amber codons within the linker separating the yellow and the blue chromoproteins. Therefore, the expression of a suppressor t-RNA will suppress amber codons allowing the translation of the green fusion chromoprotein. Conversely, the down regulation of the suppressor t-RNA through time will allow bacteria switch from green to yellow, thus simulating the ripening of a real lemon. This system will be referred to as the color switch system.
The green fusion chromoprotein design
Promoter & RBS
Because the ripening of the lemon will be achieved by the regulation of the expression of the suppressor t-RNA, the fusion chromoprotein has been placed under control of a constitutive promoter. To ensure that E. coli produces a robust color, we chose the RBS BBa_B0034 (strength = 1) and the “consensus” promoter BBa_J23119.
Chromoproteins
Uppsala iGEM team 2013 already designed several chromoproteins such as the FwYellow (BBa_K1033910) and the AeBlue chromoproteins (BBa_K1033902) [1]. We chose to fuse these chromoproteins so it should produce a green color.
Linker
Linkers are short sequences usually containing flexible amino-acid such as glycine and serine to ensure that two proteins domain are correctly folded and avoid steric issues. Thus, a linker of 12 amino acids that repeat a motif of glycine-glycine-serine-glycine (BBa_K243006) has been used to separate both FwYellow and AeBlue chromoproteins. Furthermore, to permit the color switch by the translational suppression system, we plan to replace two serine codons from the linker by amber codons (TAG). Choosing to add two amber codons would ensure a low stop codon readthrough in absence of the t-RNA.
Terminator
To end the transcription of the fusion chromoprotein, a commonly used terminator BBa_B0010 is added.
The Color Switch system design
The supD suppressor t-RNA
Given that the linker that separates both FwYellow and AeBlue chromoproteins is composed of serine and glycine amino-acid, we needed a t-RNA suppressor that could encode one of these amino-acids to not alter the properties of the linker. As we were looking for a t-RNA suppressor, we found that the iGEM Beijing 2009 team already worked on a translational suppression system. They worked with the supD suppressor t-RNA that encodes a serine making it an ideal candidate for our project. They placed supD under control of a salicylate inducible promoter Psal to suppress amber codon they had introduced in the T7 polymerase sequence. In turn, the T7 polymerase can express the GFP output gene. Their results show that supD does not induce bacteria lethality so such a system could be used [2].
Transcription factor sensible to salicylate
The nahR gene is involved in the degradation of the naphthalene pollutant in Pseudomonas putida. This gene encodes a transcriptional regulator that is induce by salicylate and thus bind nah or sal promoters. The BBa_K228004 Biobrick® contains the nahR gene under control of a constitutive promoter and the salicylate promoter (Psal). Thus, we plan to place supD under control of the Psal promoter.
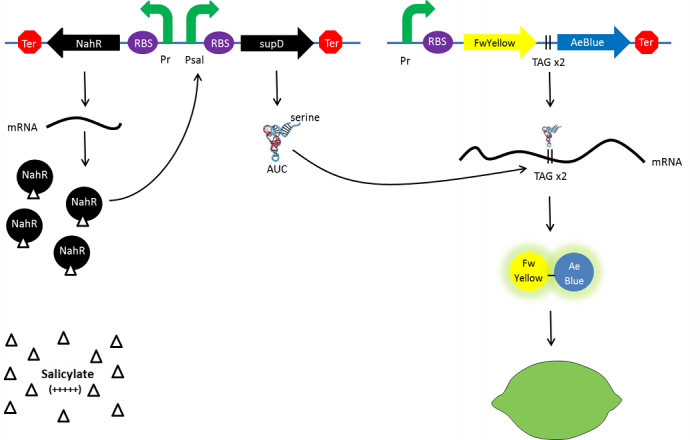
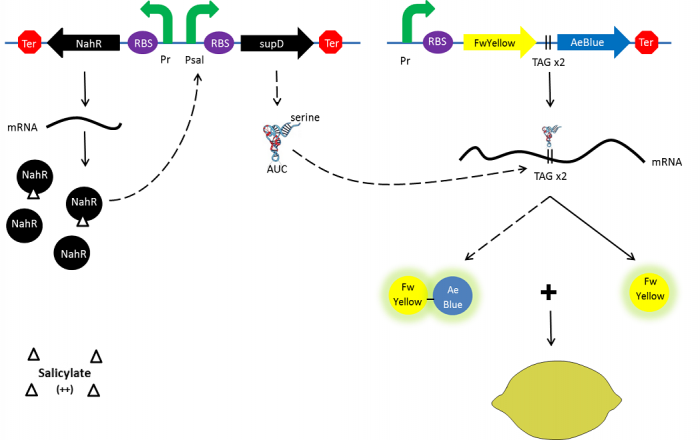
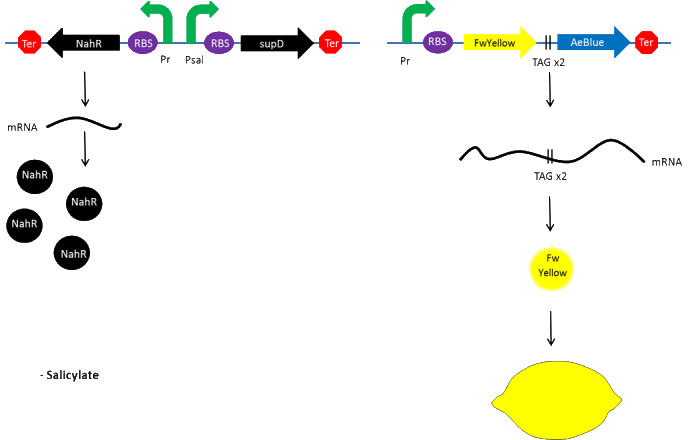
Color switch mechanism
In the beginning, salicylate concentration will be maximal into the agar media so that supD will be expressed and so the green fusion chromoprotein: bacteria will display a green color. However, as bacteria grow into agar, less salicylate will remain available into the media. Thus, the decrease of the nahR-salicylate complex amount within bacteria will lead to supD downregulation through time. In turn, decrease of supD amount will result in less codon readthrough and so less translation of the green fusion protein and more translation of the yellow chromoprotein. As a result, bacteria will gradually change from green to yellow.
Results
To save time the fusion chromoprotein gene was ordered to Lifetechnologies® company. It includes the suffix, the promoter, the RBS, the FwYellow’s open reading frame, the linker, the AeBlue’s open reading frame, the terminator and the prefix. As a first step, the linker did not bear the two amber codons so that we could know if the fusion chromoprotein is functional or not.
Cloning in pGEMTeasy
After receiving it, the fusion chromoprotein was amplified by PCR and then cloned into pGEMTeasy. Unfortunately, after transforming E.coli and plating the tranformation mix, no colonies display any color. We sequenced the gene cloned to ensure the absence of mutation. The simplest explanation is that the fusion protein is not anymore functional. One may suggest that the fact the chromoproteins need to be in a multimeric form to be functional, prevent the fusion protein to be functional.
As the fusion chromoprotein did not work, the colour switch system that we planned to do is no longer valid.
Alternative pathway
To resolve this issue, we planned to separately express both FwYellow and AeBlue in bacteria. We decided to place the FwYellow under control of a constitutive promoter whereas AeBlue will be place under the Psal inducible promoter, as mentioned above. As previously, the beginning, salicylate concentration is maximal into the agar media leading so both chromoprotein will be expressed. Assuming that the presence of both chromoprotein in the cytoplasm will display a green color, bacteria will be green. However, as bacteria grow into the agar media, the amount of nahR-salicylate complex will decrease through time. Thus, the expression of the AeBlue will decrease whereas the expression of FwYellow will remain equal. As a result, bacteria will gradually change from green to yellow.
References
[1] https://2013.igem.org/Team:Uppsala/chromoproteins
[2] https://2009.igem.org/Team:PKU_Beijing/Parts_Characterization/BBa_K228258
 "
"