Team:Freiburg/Content/Project/Vision
From 2014.igem.org
| (82 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
</head> | </head> | ||
<body> | <body> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<div class="row category-row"> | <div class="row category-row"> | ||
| - | + | <div class="col-sm-6"> | |
| - | + | <div class="container-fluid" style="float: left"> | |
| - | + | <div style="position: relative; float: right; margin-top: 4px;"> | |
| - | + | <a href="https://2014.igem.org/Team:Freiburg/Project/The_viral_vector">Go back to The Viral Vector</div> | |
| + | <div style="position: relative; float: left;"> <img class="img-no-border" style="max-width: 50px; margin-top:5px;" src=" https://static.igem.org/mediawiki/2014/4/44/Freiburg2014_Navigation_Arrow_rv.png"> <!-- Pfeil rv--></a></div> | ||
</div> | </div> | ||
| - | + | </div> | |
| - | + | <div class="col-sm-6"> | |
| - | + | <div class="container-fluid" style="float: right"> | |
| - | + | <div style="position: relative; float: left; margin-top: 4px;"> | |
| - | + | <a href="https://2014.igem.org/Team:Freiburg/Project/Outlook">Have a look at our Outlook</div> | |
| - | + | <div style="position: relative; float: right;"> <img class="img-no-border" style="max-width: 50px; margin-top:5px;" src=" https://static.igem.org/mediawiki/2014/9/95/Freibur2014_pfeilrechts.png"> <!-- Pfeil fw--></a></div> | |
| - | + | ||
</div> | </div> | ||
| - | < | + | <p> </p> |
| + | <div class="container-fluid" style="float: right"> | ||
| + | <div style="position: relative; float: left; margin-top: 4px;"> | ||
| + | |||
| + | <a href="https://2014.igem.org/Team:Freiburg/Results">Or go on to our Results</div> | ||
| + | <div style="position: relative; float: right;"> <img class="img-no-border" style="max-width: 50px; margin-top:5px;" src=" https://static.igem.org/mediawiki/2014/9/95/Freibur2014_pfeilrechts.png"> <!-- Pfeil fw--></a></div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <section id="Project-Vision"> | ||
| + | <h1>Synthetic Biology: Going from Prokaryotic to Mammalian Systems</h1> | ||
| + | |||
| + | <div class="row category-row"> | ||
| + | <div class="col-sm-6"> | ||
| + | <p> | ||
| + | During the first phase of our project we had fierce discussions about possible projects. | ||
| + | Our members collected information about current research and did some brainstorming, | ||
| + | resulting in long and interesting debates. In summary, we had a lot of fun in our | ||
| + | finding phase. Yet most of us never competed in iGEM or even heard about it before, | ||
| + | thus having no idea what iGEM is all about. Therefore, we examined previous | ||
| + | iGEM projects of other teams as an inspiration. While we analyzed a lot of these projects, | ||
| + | we noticed that by far most of the teams favor bacterial systems over mammalian cells | ||
| + | for their projects. Especially <em class="highlight-kursiv">Escherichia coli</em> and | ||
| + | <em class="highlight-kursiv">Bacillus subtilis</em> are the number one organisms in iGEM | ||
| + | history, and this year’s competition will make no difference. | ||
| + | </p> | ||
| + | <p> | ||
| + | Mammalian cells can form multicellular patterns and are of much interest in clinical applications. | ||
| + | So we asked ourselves: why are there so few iGEM teams working with mammalian cells? | ||
| + | Why do most teams prefer to work with bacterial systems? And: <em>can we develop a | ||
| + | system that facilitates dealing with mammalian cells, so that more future iGEM teams | ||
| + | consider to work with mammalian cells?</em> | ||
| + | </p> | ||
| + | </div> | ||
| + | <div class="col-sm-6"; style="text-align: center"> | ||
| + | <figure> | ||
| + | <a href="https://static.igem.org/mediawiki/2014/a/a8/Freiburg2014_Competition_2013_Organisms_sm.png"> | ||
| + | <img style="max-width: 70%"; | ||
| + | src="https://static.igem.org/mediawiki/2014/a/a8/Freiburg2014_Competition_2013_Organisms_sm.png"> | ||
| + | </a> | ||
| + | <figcaption> | ||
| + | <p class="header">Systems used at iGEM 2013.</p> | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <div class="row category-row"> | ||
| + | <div class="col-sm-6"> | ||
| + | <p> | ||
| + | There are several obvious reasons for this: First, working with mammalian cell lines | ||
| + | is normally far more expensive than working with bacteria or algae and plants. That starts | ||
| + | with medium and goes on with special equipment and labs, which often are required. Next, | ||
| + | <em class="highlight-kursiv">E. coli</em> and <em class="highlight-kursiv">B. subtilis</em> | ||
| + | are the best described, categorized and established system in the iGEM competition. Finally, | ||
| + | prokaryotic synthetic biology is right now the largest field of synthetic biology. Consequently | ||
| + | many supervisors are specialists in prokaryotic methods, thus leading to a stronger | ||
| + | focus on prokaryotic projects. | ||
| + | </p> | ||
| + | <p> | ||
| + | However, another big problem is the time consuming and often complicated handling of | ||
| + | mammalian cells. Particularly the generation of stable cell lines is a major obstacle for many teams. | ||
| + | </p> | ||
| + | </div> | ||
| + | <div class="col-sm-6"> | ||
| + | <figure> | ||
| + | <a href="https://static.igem.org/mediawiki/2014/0/00/Freiburg2014_Reasons_sm.png"> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/0/00/Freiburg2014_Reasons_sm.png"> | ||
| + | </a> | ||
| + | </figure> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <div class="row category-row"> | ||
| + | <div class="col-sm-6"> | ||
| + | <p> | ||
| + | <em>But why is the generation of stable cell lines that necessary?</em> | ||
| + | </p> | ||
| + | <p> | ||
| + | For synthetic biology, controlled and sustained expression of multiple genes is | ||
| + | usually required. In most cases, the different genes will be introduced into the target | ||
| + | cell via several plasmids. Bacteria with sustained propagation of several different plasmids | ||
| + | can be generated fast and easy, but transfections, the “normal” way to introduce plasmids | ||
| + | into mammalian cells, suffer from lack of control over expression levels and the inability | ||
| + | to introduce three or more plasmids at the same time. | ||
| + | </p> | ||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | <div class="row category-row"> | ||
| + | <div class="col-sm-6"> | ||
| + | |||
| + | <p> | ||
| + | Therefore, it is necessary for synthetic biology projects with mammalian cells to generate | ||
| + | stable cell lines, where the genes of interest are stably integrated into the genome. | ||
| + | Although this is possible with transfections and subsequent selection with antibiotics, | ||
| + | it is very time consuming because only one gene can be introduced at a time, and integration | ||
| + | events are very rare, leading to long times (weeks) of cell line expansion before the next step. | ||
| + | </p> | ||
| + | <p> | ||
| + | The solution to this problem is the generation of stable cell lines via viral transduction, | ||
| + | where the gene of interest is stably integrated into the host genome. Due to the high efficiency | ||
| + | of this process, the cells can be sorted very soon after gene transfer, and the resulting stable | ||
| + | cell line is ready for the next step after only one week. In addition, the high efficiency | ||
| + | of gene transfer makes the simultaneous introduction of several genes within a single step possible. | ||
| + | </p> | ||
| + | <p> | ||
| + | The most common viral system for transduction is the lentiviral system. However, due to | ||
| + | its promiscuity to infect mammalian cells of many species (including humans), working with | ||
| + | lentivirus requires an S2 biosafety standard. For some iGEM teams this may be just an | ||
| + | additional inconvenience, but for others it can mean the end of the project. | ||
| + | </p> | ||
| + | </div> | ||
| + | <div class="col-sm-6"; style="text-align:center"> | ||
| + | <figure> | ||
| + | <a href="https://static.igem.org/mediawiki/2014/2/26/Freiburg2014_Cell_line_sm.png"> | ||
| + | <img style="max-width: 70%"; | ||
| + | src="https://static.igem.org/mediawiki/2014/2/26/Freiburg2014_Cell_line_sm.png"> | ||
| + | </a> | ||
| + | </figure> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <div class="row category-row"> | ||
| + | <div class="col-sm-6"> | ||
| + | <p> | ||
| + | To overcome these limitations, our team searched for an alternative to the lentivirus | ||
| + | that can be used under S1 biosafety standard. The requirements for this virus are: | ||
| + | easy preparation, large cargo, high transduction efficiency, but a very high specificity | ||
| + | towards a defined non-human cell type. After consulting with virus specialists we found | ||
| + | a virus that perfectly suited our demands: the Murine Leukemia Virus (MuLV). | ||
| + | </p> | ||
| + | <p> | ||
| + | <em> | ||
| + | The MuLV offers the following great advantages: | ||
| + | <ul type="disk"> | ||
| + | <li>only infects murine cells</li> | ||
| + | <li>carries up to 8 kB of DNA</li> | ||
| + | <li>easy generation in Phoenix cells</li> | ||
| + | <li>high transduction efficiency</li> | ||
| + | <li>not patented</li> | ||
| + | <li>can be used with non-murine cells (!) after transfection of a single protein</li> | ||
| + | </ul> | ||
| + | </em> | ||
| + | </p> | ||
| + | <p> | ||
| + | We, the iGEM team Freiburg 2014, established the MuLV for the iGEM community. | ||
| + | We optimized the protocols for working with the MuLV and indeed generated stable cell | ||
| + | lines with several genes. For use by other teams, | ||
| + | the Phoenix cell line can be obtained by ATCC, and the plasmid for virus production | ||
| + | has been deposited as a backbone to the iGEM repository. Have fun cloning! | ||
| + | </p> | ||
| + | |||
| + | </div> | ||
| + | <div class="col-sm-6"; style="text-align:center"> | ||
| + | <figure> | ||
| + | <a href="https://static.igem.org/mediawiki/2014/e/e6/Freiburg2014_MuLV.png"> | ||
| + | <img style="max-width: 60%"; src="https://static.igem.org/mediawiki/2014/e/e6/Freiburg2014_MuLV.png"> | ||
| + | </a> | ||
| + | <figcaption> | ||
| + | <p class="header">Properties of the Murine Leukemia Virus.</p> | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | </section> | ||
| + | <div class="row category-row"> | ||
| + | <div class="col-sm-6"> | ||
| + | <div class="container-fluid" style="float: left"> | ||
| + | <div style="position: relative; float: right; margin-top: 4px;"> | ||
| + | <a href="https://2014.igem.org/Team:Freiburg/Project/The_viral_vector">Go back to The Viral Vector</div> | ||
| + | <div style="position: relative; float: left;"> <img class="img-no-border" style="max-width: 50px; margin-top:5px;" src=" https://static.igem.org/mediawiki/2014/4/44/Freiburg2014_Navigation_Arrow_rv.png"> <!-- Pfeil rv--></a></div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="col-sm-6"> | ||
| + | <div class="container-fluid" style="float: right"> | ||
| + | <div style="position: relative; float: left; margin-top: 4px;"> | ||
| + | <a href="https://2014.igem.org/Team:Freiburg/Project/Outlook">Have a look at our Outlook</div> | ||
| + | <div style="position: relative; float: right;"> <img class="img-no-border" style="max-width: 50px; margin-top:5px;" src=" https://static.igem.org/mediawiki/2014/9/95/Freibur2014_pfeilrechts.png"> <!-- Pfeil fw--></a></div> | ||
| + | </div> | ||
| + | <p> </p> | ||
| + | <div class="container-fluid" style="float: right"> | ||
| + | <div style="position: relative; float: left; margin-top: 4px;"> | ||
| + | |||
| + | <a href="https://2014.igem.org/Team:Freiburg/Results">Or go on to our Results</div> | ||
| + | <div style="position: relative; float: right;"> <img class="img-no-border" style="max-width: 50px; margin-top:5px;" src=" https://static.igem.org/mediawiki/2014/9/95/Freibur2014_pfeilrechts.png"> <!-- Pfeil fw--></a></div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
</body> | </body> | ||
</html> | </html> | ||
Latest revision as of 02:50, 18 October 2014
Synthetic Biology: Going from Prokaryotic to Mammalian Systems
During the first phase of our project we had fierce discussions about possible projects. Our members collected information about current research and did some brainstorming, resulting in long and interesting debates. In summary, we had a lot of fun in our finding phase. Yet most of us never competed in iGEM or even heard about it before, thus having no idea what iGEM is all about. Therefore, we examined previous iGEM projects of other teams as an inspiration. While we analyzed a lot of these projects, we noticed that by far most of the teams favor bacterial systems over mammalian cells for their projects. Especially Escherichia coli and Bacillus subtilis are the number one organisms in iGEM history, and this year’s competition will make no difference.
Mammalian cells can form multicellular patterns and are of much interest in clinical applications. So we asked ourselves: why are there so few iGEM teams working with mammalian cells? Why do most teams prefer to work with bacterial systems? And: can we develop a system that facilitates dealing with mammalian cells, so that more future iGEM teams consider to work with mammalian cells?
There are several obvious reasons for this: First, working with mammalian cell lines is normally far more expensive than working with bacteria or algae and plants. That starts with medium and goes on with special equipment and labs, which often are required. Next, E. coli and B. subtilis are the best described, categorized and established system in the iGEM competition. Finally, prokaryotic synthetic biology is right now the largest field of synthetic biology. Consequently many supervisors are specialists in prokaryotic methods, thus leading to a stronger focus on prokaryotic projects.
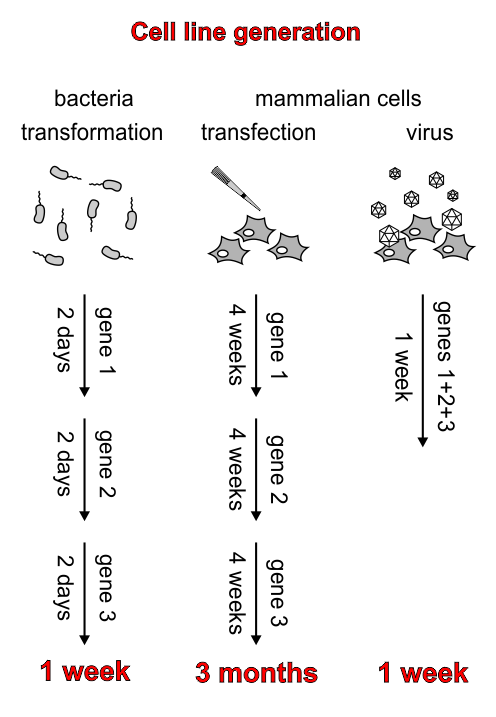
However, another big problem is the time consuming and often complicated handling of mammalian cells. Particularly the generation of stable cell lines is a major obstacle for many teams.
But why is the generation of stable cell lines that necessary?
For synthetic biology, controlled and sustained expression of multiple genes is usually required. In most cases, the different genes will be introduced into the target cell via several plasmids. Bacteria with sustained propagation of several different plasmids can be generated fast and easy, but transfections, the “normal” way to introduce plasmids into mammalian cells, suffer from lack of control over expression levels and the inability to introduce three or more plasmids at the same time.
Therefore, it is necessary for synthetic biology projects with mammalian cells to generate stable cell lines, where the genes of interest are stably integrated into the genome. Although this is possible with transfections and subsequent selection with antibiotics, it is very time consuming because only one gene can be introduced at a time, and integration events are very rare, leading to long times (weeks) of cell line expansion before the next step.
The solution to this problem is the generation of stable cell lines via viral transduction, where the gene of interest is stably integrated into the host genome. Due to the high efficiency of this process, the cells can be sorted very soon after gene transfer, and the resulting stable cell line is ready for the next step after only one week. In addition, the high efficiency of gene transfer makes the simultaneous introduction of several genes within a single step possible.
The most common viral system for transduction is the lentiviral system. However, due to its promiscuity to infect mammalian cells of many species (including humans), working with lentivirus requires an S2 biosafety standard. For some iGEM teams this may be just an additional inconvenience, but for others it can mean the end of the project.
To overcome these limitations, our team searched for an alternative to the lentivirus that can be used under S1 biosafety standard. The requirements for this virus are: easy preparation, large cargo, high transduction efficiency, but a very high specificity towards a defined non-human cell type. After consulting with virus specialists we found a virus that perfectly suited our demands: the Murine Leukemia Virus (MuLV).
The MuLV offers the following great advantages:
We, the iGEM team Freiburg 2014, established the MuLV for the iGEM community. We optimized the protocols for working with the MuLV and indeed generated stable cell lines with several genes. For use by other teams, the Phoenix cell line can be obtained by ATCC, and the plasmid for virus production has been deposited as a backbone to the iGEM repository. Have fun cloning!
 "
"