Team:Freiburg/Content/Project/Overview
From 2014.igem.org
| (117 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | <!-- hier ändern https://2014.igem.org/Team:Freiburg/Content/Project/Overview --> | + | <!-- hier ändern https://2014.igem.org/Team:Freiburg/Content/Project/Project-Overview --> |
| - | <html> | + | <html lang="en"> |
| + | <head> | ||
| + | <title>The AcCELLerator</title> | ||
| + | <link rel="stylesheet" type="text/css" href="styles.css"> | ||
| + | </head> | ||
| + | <body> | ||
| - | <section id="Overview"> | + | <section id="Project-Overview-Vision"><!-- zu bootstrap hinzufügen--> |
| + | <blockquote> | ||
| + | <h1><strong>Summary</strong></h1> <!-- zu bootstrap hinzufügen--> | ||
| + | |||
| + | <p>The rapid development of synthetic biology is accompanied by an increased demand for methods that should not only simplify gene delivery, but also provide spatial and temporal control over gene expression. | ||
| + | We, the iGEM team Freiburg 2014, designed a system, which combines the advantages of two biotechnological approaches – the spatio-temporal resolution of optogenetics and the simplicity of viral gene transfer. In our engineered cell line, illumination with light of a defined wavelength leads to the expression of a specific receptor. This receptor serves as an entry point for a viral vector carrying the genes to be inserted. This combined approach provides a comprehensive picture of the cellular output resulting from different genetic inputs. The gained knowledge can be used to monitor processes of cell development in a new dimension of complexity and ultimately enable facilitated tissue engineering. | ||
| + | </p> | ||
| + | <a href="https://2014.igem.org/Team:Freiburg/Project/Vision" class="read-more">Read the story behind <strong>The AcCELLerator</strong></a> | ||
| + | |||
| + | </blockquote> | ||
| + | </section> | ||
| + | |||
| + | |||
| + | <section id="Project-Overview"> | ||
<h1>Overview</h1> | <h1>Overview</h1> | ||
| - | <h2 id="Overview-Introduction">Introduction</h2> | + | <h2 id="Project-Overview-Introduction">Introduction</h2> |
<div class="row category-row"> | <div class="row category-row"> | ||
<div class="col-sm-6"> | <div class="col-sm-6"> | ||
| - | <p>Gene expression is one of the most complex and tightly controlled processes within eukaryotic cells. Cellular | + | <p>Gene expression is one of the most complex and tightly controlled processes within eukaryotic cells. Cellular processes crucially depend on precise coordination of multiple genes.</p> |
| - | <p> | + | <p>In order to study these processes, viral vectors can be used to efficiently deliver diverse genetic elements for regulating gene expression in cells. These can include transcription factors, genome editing devices such as the CRISPR system, or other genes of choice. However, often spatio-temporally controlled introduction of genetic elements is necessary, which is not provided by viral vectors.</p> |
| + | <p>Optogenetic tools such as light-responsive gene regulation systems have become more and more prominent over the last few years, enabling high spatio-temporal control over gene expression. </p> | ||
| + | <br> | ||
| + | |||
| + | <p>For the past six months our team developed a novel application, called <strong>The AcCELLerator</strong>, which combines the advantages of the above mentioned techniques. We use a blue light-inducible gene expression system to spatio-temporally control the cargo delivery via viral vectors. This combination facilitates the delivery of variable exogenous genes and provides control over endogenous genes, both with high spatio-temporal resolution.</p> | ||
| + | <p>In addition, The AcCELLerator enables the fast and easy generation of stable cell lines. Stable cell lines are required for constant and predictable gene expression, as transient introduction of target genes entails the risk of unstable and discontinuous expression levels. To date, viral vectors are commonly used for the generation of stable cell lines. However, most of them demand work under biosafety level 2 conditions. The AcCELLerator, based on a biosafety level 1 viral vector, simplifies the existing time-consuming methods without the need of special safety precautions. Thereby it makes mammalian research <a href="https://2014.igem.org/Team:Freiburg/Project/Vision" class="read-more">more attractive for the iGEM community.</a></p> | ||
| + | |||
</div> | </div> | ||
<div class="col-sm-6"> | <div class="col-sm-6"> | ||
| - | + | <figure> | |
| + | <a href="https://static.igem.org/mediawiki/2014/b/b1/Freiburg2014_schema_new_figure1_project.png"> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/b/b1/Freiburg2014_schema_new_figure1_project.png"> | ||
| + | </a> | ||
| + | |||
| + | </figure> | ||
</div> | </div> | ||
</div> | </div> | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</section> | </section> | ||
<section> | <section> | ||
| - | <h2 id="Overview-Principle"> | + | <h2 id="Project-Overview-Principle">The AcCELLarator - Optogenetic Control of Viral Gene Delivery</h2> |
<div class="row category-row"> | <div class="row category-row"> | ||
<div class="col-sm-6"> | <div class="col-sm-6"> | ||
| - | + | <figure> | |
| + | </html>{{:Team:Freiburg/svg/Principle.svg}}<html> | ||
| + | |||
| + | </figure> | ||
</div> | </div> | ||
<div class="col-sm-6"> | <div class="col-sm-6"> | ||
| - | <p> | + | <p>Our tool is a two-component system, combing specificity of optogenetics with variability of viral vectors. The light system is stably integrated into a cell line. It controls the expression of a certain receptor. When illuminated with blue light, the light system switches into the “ON”-state and the receptor is expressed. Cells expressing the receptor can be recognized by our second component, a viral vector. The DNA cargo of the viral vector is easily exchangeable.</p> |
</div> | </div> | ||
</div> | </div> | ||
</section> | </section> | ||
<section> | <section> | ||
| - | <h2 id="Overview-Light-System">Light | + | <h2 id="Project-Overview-Light-System">Light System</h2> |
<div class="row category-row"> | <div class="row category-row"> | ||
<div class="col-sm-6"> | <div class="col-sm-6"> | ||
| - | <p> | + | <p>Gene expression systems under the control of light are based on two components: One part is constantly located at a specific promoter-associated DNA sequence, the second is fused to a transactivator domain. The system can be in two different states: ON and OFF. In the dark, the system is in the OFF state. Upon illumination, the system is switched ON. The light-induced interaction of both units leads to the recruitment of a DNA polymerase and henceforth the target gene is expressed.<br> |
| - | + | <a href="https://2014.igem.org/Team:Freiburg/Project/The_light_system">Read More about the Light System</a></p> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
| - | + | <div class="col-sm-6"> | |
| - | + | ||
<figure> | <figure> | ||
| - | + | <a href="https://static.igem.org/mediawiki/2014/5/55/2014Freiburg_Lov2system.gif"> | |
| - | + | <img src="https://static.igem.org/mediawiki/2014/3/35/2014FreiburgLight-system-thumbnail.gif"> | |
| - | + | </a> | |
| - | + | ||
| - | + | </figure> | |
| - | + | ||
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</section> | </section> | ||
<section> | <section> | ||
| - | <h2 id="Overview-Viral-Vectors">Viral | + | <h2 id="Project-Overview-Viral-Vectors">Viral Vectors And The Receptor</h2> |
<div class="row category-row"> | <div class="row category-row"> | ||
<div class="col-sm-6"> | <div class="col-sm-6"> | ||
| - | <p> | + | <p>By using viral vectors it is possible to deliver and stably integrate a specific gene cargo within only four days. As the viral recognition site, a specific receptor represents the key component of viral gene delivery. Only if the receptor is present on the cell surface, the virus is able to insert the gene of interest. Otherwise, the virus cannot enter the cells. <br> |
| + | <a href="https://2014.igem.org/Team:Freiburg/Project/The_viral_vector">Read More about Viral Vectors</a> and <a href="https://2014.igem.org/Team:Freiburg/Project/Receptor">the Receptor</a></p> | ||
| + | |||
| + | </div> | ||
<div class="col-sm-6"> | <div class="col-sm-6"> | ||
| - | <figure> | + | <figure style="display:inline-block;text-align:center;"> |
| - | + | <a href="https://static.igem.org/mediawiki/2014/7/75/Freiburg2014_schema_new_figure3_project.png"> | |
| - | + | <img style="max-width: 400px;" src="https://static.igem.org/mediawiki/2014/7/75/Freiburg2014_schema_new_figure3_project.png"> | |
| - | + | </a> | |
<figcaption> | <figcaption> | ||
| - | <p class="header"> | + | <p class="header"></p> |
</figcaption> | </figcaption> | ||
</figure> | </figure> | ||
| Line 98: | Line 103: | ||
</div> | </div> | ||
</section> | </section> | ||
| - | + | <div class="row category-row"> | |
| - | + | <div class="col-sm-6"> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
| - | < | + | <div class="col-sm-6"> |
| - | + | <div class="container-fluid" style="float: right"> | |
| - | + | <div style="position: relative; float: left; margin-top: 4px;"> | |
| - | + | <a href="https://2014.igem.org/Team:Freiburg/Project/The_light_system">Start our Tour and Read more about the Light System</div> | |
| - | + | <div style="position: relative; float: right;"> <img class="img-no-border" style="max-width: 50px; margin-top:5px;" src=" https://static.igem.org/mediawiki/2014/9/95/Freibur2014_pfeilrechts.png"> <!-- Pfeil fw--></a></div> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| + | </div> | ||
| - | + | </body> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | </ | + | |
| - | + | ||
</html> | </html> | ||
Latest revision as of 03:23, 18 October 2014
Summary
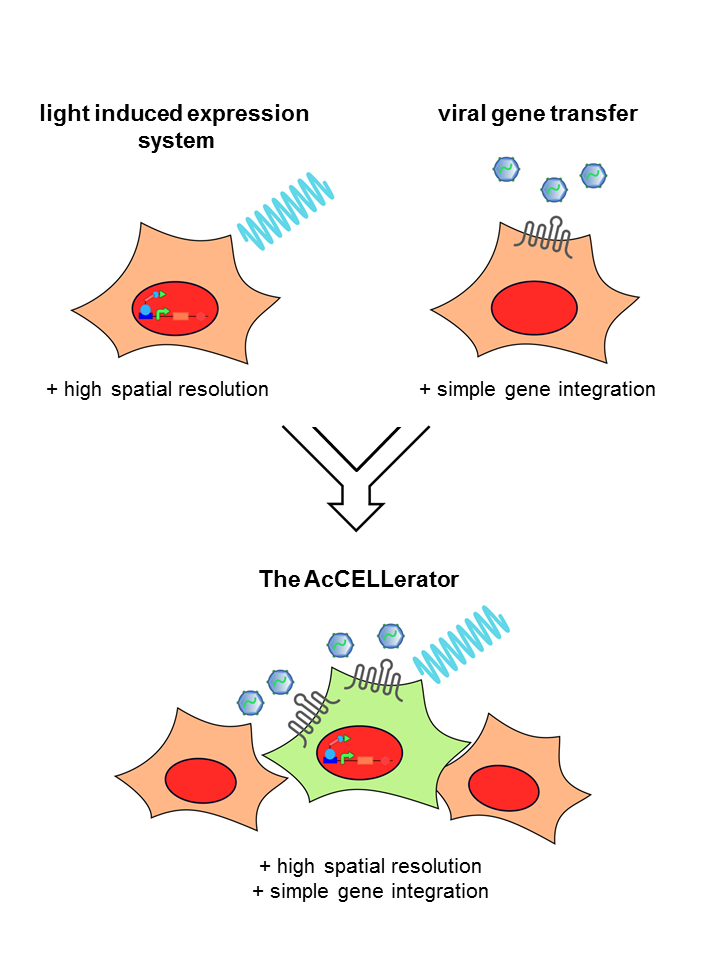
The rapid development of synthetic biology is accompanied by an increased demand for methods that should not only simplify gene delivery, but also provide spatial and temporal control over gene expression. We, the iGEM team Freiburg 2014, designed a system, which combines the advantages of two biotechnological approaches – the spatio-temporal resolution of optogenetics and the simplicity of viral gene transfer. In our engineered cell line, illumination with light of a defined wavelength leads to the expression of a specific receptor. This receptor serves as an entry point for a viral vector carrying the genes to be inserted. This combined approach provides a comprehensive picture of the cellular output resulting from different genetic inputs. The gained knowledge can be used to monitor processes of cell development in a new dimension of complexity and ultimately enable facilitated tissue engineering.
Read the story behind The AcCELLerator
Overview
Introduction
Gene expression is one of the most complex and tightly controlled processes within eukaryotic cells. Cellular processes crucially depend on precise coordination of multiple genes.
In order to study these processes, viral vectors can be used to efficiently deliver diverse genetic elements for regulating gene expression in cells. These can include transcription factors, genome editing devices such as the CRISPR system, or other genes of choice. However, often spatio-temporally controlled introduction of genetic elements is necessary, which is not provided by viral vectors.
Optogenetic tools such as light-responsive gene regulation systems have become more and more prominent over the last few years, enabling high spatio-temporal control over gene expression.
For the past six months our team developed a novel application, called The AcCELLerator, which combines the advantages of the above mentioned techniques. We use a blue light-inducible gene expression system to spatio-temporally control the cargo delivery via viral vectors. This combination facilitates the delivery of variable exogenous genes and provides control over endogenous genes, both with high spatio-temporal resolution.
In addition, The AcCELLerator enables the fast and easy generation of stable cell lines. Stable cell lines are required for constant and predictable gene expression, as transient introduction of target genes entails the risk of unstable and discontinuous expression levels. To date, viral vectors are commonly used for the generation of stable cell lines. However, most of them demand work under biosafety level 2 conditions. The AcCELLerator, based on a biosafety level 1 viral vector, simplifies the existing time-consuming methods without the need of special safety precautions. Thereby it makes mammalian research more attractive for the iGEM community.
The AcCELLarator - Optogenetic Control of Viral Gene Delivery
Our tool is a two-component system, combing specificity of optogenetics with variability of viral vectors. The light system is stably integrated into a cell line. It controls the expression of a certain receptor. When illuminated with blue light, the light system switches into the “ON”-state and the receptor is expressed. Cells expressing the receptor can be recognized by our second component, a viral vector. The DNA cargo of the viral vector is easily exchangeable.
Light System
Gene expression systems under the control of light are based on two components: One part is constantly located at a specific promoter-associated DNA sequence, the second is fused to a transactivator domain. The system can be in two different states: ON and OFF. In the dark, the system is in the OFF state. Upon illumination, the system is switched ON. The light-induced interaction of both units leads to the recruitment of a DNA polymerase and henceforth the target gene is expressed.
Read More about the Light System
Viral Vectors And The Receptor
By using viral vectors it is possible to deliver and stably integrate a specific gene cargo within only four days. As the viral recognition site, a specific receptor represents the key component of viral gene delivery. Only if the receptor is present on the cell surface, the virus is able to insert the gene of interest. Otherwise, the virus cannot enter the cells.
Read More about Viral Vectors and the Receptor
 "
"