Team:BIOSINT Mexico/Chassis
From 2014.igem.org
| (49 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:BIOSINT_Mexico/Header2}} | {{:Team:BIOSINT_Mexico/Header2}} | ||
| - | + | [[File:Chassis.png|800px|center]] | |
| - | + | ||
| - | + | <html><h1>Arabidopsis - standar chassis in iGEM</h1> </html> | |
| - | + | <html><h2>Description</h2> </html> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | . | + | |
| - | + | In biotechnology plants are mainly used for premeditation, for the obtainment of industrial products and to generate energy. For these reasons plant manipulation is a focus area in biological engineering. Unfortunately there is not a wide variety of projects in iGEM using plants as a chassis and one of the main reasons is the lack of information in the registry of these organisms. | |
| - | + | ||
| - | + | Compared with other organisms used as models, the proportion of ''A. thaliana'' proteins have related counterparts in Eukaryota genomes, these varies by a factor of 2 to 3, depending on the functional category. Only 8-23% of ''A. thaliana'' proteins involved in transcription have related genes in other Eukaryota genomes, reflecting the independent evolution of many plant transcription factors. | |
| - | + | ||
| - | + | In contrast, 48-60% of genes involved in protein synthesis have counterpart in other eukaryota genomes, reflecting highly conserved gene functions. The relatively high proportion of matches between ''A. thaliana'' and bacterial proteins in the categories “metabolism” and “energy” reflects, both, the acquisition of bacterial genes from the ancestor of the plasmid and high conservation of sequences across all species. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | ''Arabidopsis thaliana'' is a model organism that was previously described as chassis. In iGEM 2010, Harvard team used this plant to test their designed vectors for ''Agrobacterium tumefaciens''; in 2012 the Kyoto team work with ''A. thaliana'' as model for the induction of flower formation by putting ''E. coli'' in the leaves. |
| - | + | ||
| - | + | <font size="3">[[File:Harv.png|500px|thumb|center|'''Figure 1''' iGarden by Harvard 2010 iGEM team.]]</font> | |
| - | + | ||
| - | + | ||
| - | + | Harvard developed six ''Agrobacterium '' vectors to mediated plant transformation. These were modified from the pORE system to fit in the standard Biobrick, using PCR mutagenesis and digestion to replace the multiple cloning site with the Biobrick cloning site. The plasmids have plant resistance to kanamycin as a marker, these also contained a reporters, gusA and smgfp, on the end of the multiple clonning site and its expression follows the inserted construct. Some of the vectors contained a constitutive promoter to expressed easily the activation of the inserted genes. | |
| - | + | ||
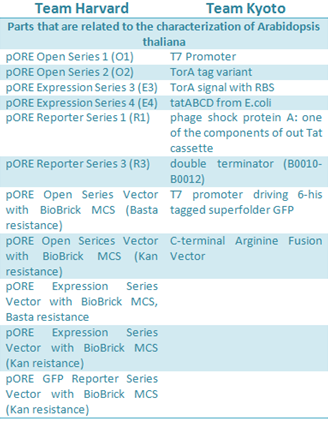
| - | + | :::<font size="3">[[File:BIOSINTpartsARA1.png|380px|thumb|left|'''Figure 2''' Parts for ''A. thaliana'' available in the registry.]]</font> | |
| - | + | ||
| - | + | ||
| - | + | ||
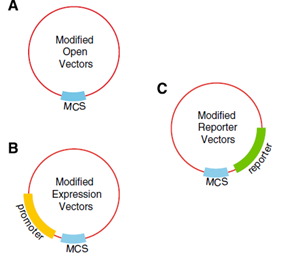
| - | + | :::::::::::<font size="3">[[File:vect.png|700px|thumb|left|'''Figure 3''' Plant Transfections Vectors.]]</font> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | </ | + | |
| - | </ | + | |
| - | </ | + | |
| - | </html> | + | |
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ===Advantages=== | ||
| + | <font size="3">[[File:Ara.JPG|500px|thumb|center|'''Figure 1''' Growing Arabidopsis Thaliana .]]</font> | ||
| + | |||
| + | *''Arabidopsis Thaliana'' is one of the first eukaryotic experimental model organism. | ||
| + | *The knowledge recollect in this plant can be applied in others organism as animals and eukaryota. | ||
| + | *It allows working with any part of the plant, seeds, leaves, root, flowers. | ||
| + | *It has a short life that make it easy to laboratory work. | ||
| + | *The self-reproduction makes it possible to obtain large population of seedlings for specific characteristic or phenotype. | ||
| + | *Small genome (114.5 Mb/125 Mb total) has been sequenced in the year 2000. | ||
| + | *Genetic and physical maps of all 5 chromosomes. | ||
| + | *A rapid life cycle (about 6 weeks from germination to mature seed). | ||
| + | *Prolific seed production and easy cultivation in restricted space. | ||
| + | *Efficient transformation methods utilizing ''Agrobacterium tumefaciens''. | ||
| + | *A large number of mutant lines and genomic resources many of which are available from Stock Centers. | ||
| + | *Multinational research community of academic, government and industry laboratories. | ||
| + | |||
| + | ===Disadvantages=== | ||
| + | |||
| + | *Does not growth faster as bacteria. | ||
| + | *There are not many parts available for plants in the registry. | ||
| + | *Transforming with ''Agrobacterium tumefaciens'' T-DNA integrates less randomly into the plant genome. | ||
| + | *High probabilities of pollution in the medium. | ||
| + | |||
| + | ===How to use Arabidopsis thaliana?=== | ||
| + | |||
| + | Successful germination and plant growth requires appropriate soil moisture, nutrient levels, light intensity, humidity, and temperature. If any of these are compromised,'' A. thaliana'' will respond by flowering early and dying prematurely and producing little leaf mass for experiments. The plants can also be stressed by overcrowding, fungus infestation, or insect infestation. All these factors must be conscientiously monitored to ensure reliable data obtained from experiments using these plants. Maintenance of soil moisture is imperative for successful seed germination. | ||
| + | |||
| + | The optimal temperature for plant growth is 25°C, with lower temperatures being allowable; higher temperatures can be very detrimental, particularly in the first two weeks of growth. | ||
| + | |||
| + | Growth conditions | ||
| + | |||
| + | With the purpose to obtain several perfects A. thaliana, the Arabidopsis Biological Resource Center (ABCR) has led methods to cultivate and grow these plants inside and outside the laboratory, in medium and soil. These methods and required conditions can be checked in the protocol “Handling Arabidopsis plants and seeds”. The growth and develop of A. thaliana depends of several environmental conditions, besides to the genetic background; with the correct conditions,light, temperature and watering, plants produce flowers within 4-5 weeks and seeds can be harvested 8 to 10 weeks after planting. | ||
| + | |||
| + | <blockquote><font size="3">[[File:BIOSINTsemi.png|330px|thumb|right|'''Figure 4''' Germination of ''A. thaliana''. ]]</font></blockquote> | ||
| + | |||
| + | With respect to the light, the optimum intensity is 120-150 umol/m2sec; higher intensities may produce death of the seedlings, while low light intensities may produce weak plants without enough chlorophyta. Besides, under photoperiods greater than 12 hours plants flower rapidly. | ||
| + | |||
| + | According to the temperature, 22-23°C ir the optimum condition, lower temperatures can be accepted but higher are impossible. The consequences with higher temperatures may result with the reduce number of the leaves, flowers and seeds while in lower temperatures growth is slow. | ||
| + | |||
| + | Other condition is the water required, the optimal humidity is the mild (50% to 60%). Lower humidity cause drying soil and higher humidity cause plant sterility. | ||
| + | |||
| + | For transformation we used the floral dip method described in this protocol: (Sushanta, 2013). | ||
| + | |||
| + | *1. Preparation of ''Agrobacterium'' strain containing the gene. Inoculate in to a 5 ml LB medium, incubate at 28°C for 2 days. | ||
| + | *2. Inoculation of culture in a 500 ml LB medium containing antibiotics. | ||
| + | *3. Centrifugate at 4000rpm for 10 min. at room temperature. | ||
| + | *4. Resuspended in 1 volume of 5% sucrose solution. 0.02% silwet L-77 is use in the mixing solution and ''Agrobacterium'' cell suspension transfer to a 500 ml beaker. ''Agrobacterium'' suspension quantity up to 400-500 ml required for transformation of at least six pots of ''A. thaliana''. | ||
| + | *5. ''Arabidopsis thaliana'' plants are invert and dip into the ''Agrobacterium'' suspension for 10 seconds with gentle agitation. | ||
| + | *6. The plants are removed from the solution and wash. | ||
| + | *7. Plants are wrapping with plastic cover, are laydown on their sides for 16-24 hours to maintain high humidity. | ||
| + | *8. On the next day, remove the cover and keep it in a growth chamber. | ||
| + | *9. Plant dry seeds collect using a sieve mesh. | ||
| + | |||
| + | <html><h1>Transformation</h1></html> | ||
| + | |||
| + | The transformation of the plant was made using V9(BBa_k382002) without the construct, this was made by using the floral deeping protocol, this was using an agrobacterium culture transformed with the vector, the flowers of the plant were placed in the medium with agrobacterium and then let it rest. | ||
| + | |||
| + | The plant were placed in a room at 21 degrees but we werent able to prove if the vector was being expressed or not. | ||
| + | |||
| + | |||
| + | <html><h2>Modeling</h2> </html> | ||
| + | |||
| + | In 2010 Harvard's 2010 iGarden used its V10 for ''Agrobacterium'' transformation and confirmed its presence with kanamycin. This team showed their results for V9 (BBa_K382002) and V10 (BBa_K382003). In their results the vectors were transformed into ''Arabidopsis'', after ''Agrobacterium'' mediated transformation, and confirmed their presence with kanamycin. | ||
| + | |||
| + | :<blockquote><font size="3">[[File:Plar.png|530px|thumb|left|'''Figure 5''' ''A. thaliana'' expressing Kanamycin resistance.]]</font></blockquote> | ||
| + | <br><br><br><br> | ||
| + | Because plants take a long time to grow, the function of the parts were unverifiable in ''Arabidopsis''. However ''E. Coli'' was used to confirm the transcription and translation of the proteins of interest. | ||
| + | |||
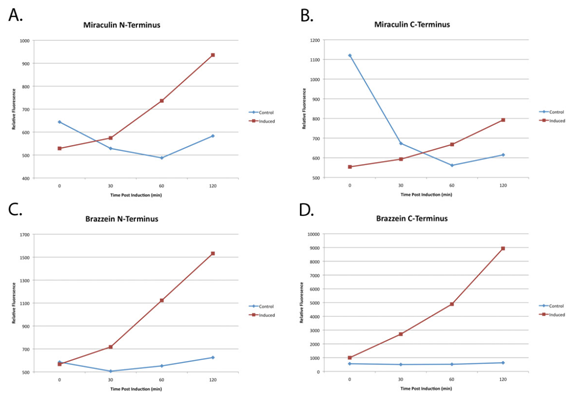
| + | ::::<blockquote><font size="3">[[File:Graf.png|660px|thumb|left|'''Figure 6''' Induced expression of YFP-tagged Miraculin and Brazzein in ''Escherichia coli''.]]</font></blockquote> | ||
| + | |||
| + | |||
| + | |||
| + | <br><br><br><br><br><br><br><br><br> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <br><br><br><br><br> | ||
| + | |||
| + | |||
| + | |||
| + | (A) through (D) are normalized plots of miraculin and brazzein YFP-fused constructs expressed in E. Coli. 2xYFP tags were attached to either the N- or C- terminus to ensure that folding was not hindered. In all cases relative YFP fluorescence had appreciably increased after 120 minutes as compared to the non-induced E. Coli | ||
| + | |||
| + | |||
| + | <html><h2>References</h2> </html> | ||
| + | *Highered.mheducation.com, (2014). Genetic Portrait Chapters A-E. [online] Available at: http://highered.mheducation.com/sites/007352526x/student_view0/genetic_portrait_chapters_a-e.html [Accessed 26 Sep. 2014]. | ||
| + | *Boyle et al.: A BrioBrick compatible strategy for genetic modification of plants. Journal of Biological Engineering 2012 6:8. | ||
| + | *Sushanta, K. (2013). Floral dip: A simple and efficient agrobacterium mediated transformation method is used in a model plant Arabidopsis Thaliana. Cibtech Journal of Bio-Protocols, 14-20. | ||
| + | *Arabidopsis Biological Resource Center, (2014). Handling Arabidopsis plants and seeds. [online] Available at: https://abrc.osu.edu/sites/abrc.osu.edu/files/abrc_handling_seed_2013.pdf [Accessed 27 Sep. 2014]. | ||
| + | *Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. (n.d.). Proquest. Retrieved October 12, 2014, from http://0-search.proquest.com.millenium.itesm.mx/docview/204469095/43FC9248199F47A8PQ/11?accountid=11643 | ||
Latest revision as of 03:59, 18 October 2014
Arabidopsis - standar chassis in iGEM
Description
In biotechnology plants are mainly used for premeditation, for the obtainment of industrial products and to generate energy. For these reasons plant manipulation is a focus area in biological engineering. Unfortunately there is not a wide variety of projects in iGEM using plants as a chassis and one of the main reasons is the lack of information in the registry of these organisms.
Compared with other organisms used as models, the proportion of A. thaliana proteins have related counterparts in Eukaryota genomes, these varies by a factor of 2 to 3, depending on the functional category. Only 8-23% of A. thaliana proteins involved in transcription have related genes in other Eukaryota genomes, reflecting the independent evolution of many plant transcription factors.
In contrast, 48-60% of genes involved in protein synthesis have counterpart in other eukaryota genomes, reflecting highly conserved gene functions. The relatively high proportion of matches between A. thaliana and bacterial proteins in the categories “metabolism” and “energy” reflects, both, the acquisition of bacterial genes from the ancestor of the plasmid and high conservation of sequences across all species.
Arabidopsis thaliana is a model organism that was previously described as chassis. In iGEM 2010, Harvard team used this plant to test their designed vectors for Agrobacterium tumefaciens; in 2012 the Kyoto team work with A. thaliana as model for the induction of flower formation by putting E. coli in the leaves.
Harvard developed six Agrobacterium vectors to mediated plant transformation. These were modified from the pORE system to fit in the standard Biobrick, using PCR mutagenesis and digestion to replace the multiple cloning site with the Biobrick cloning site. The plasmids have plant resistance to kanamycin as a marker, these also contained a reporters, gusA and smgfp, on the end of the multiple clonning site and its expression follows the inserted construct. Some of the vectors contained a constitutive promoter to expressed easily the activation of the inserted genes.
Advantages
- Arabidopsis Thaliana is one of the first eukaryotic experimental model organism.
- The knowledge recollect in this plant can be applied in others organism as animals and eukaryota.
- It allows working with any part of the plant, seeds, leaves, root, flowers.
- It has a short life that make it easy to laboratory work.
- The self-reproduction makes it possible to obtain large population of seedlings for specific characteristic or phenotype.
- Small genome (114.5 Mb/125 Mb total) has been sequenced in the year 2000.
- Genetic and physical maps of all 5 chromosomes.
- A rapid life cycle (about 6 weeks from germination to mature seed).
- Prolific seed production and easy cultivation in restricted space.
- Efficient transformation methods utilizing Agrobacterium tumefaciens.
- A large number of mutant lines and genomic resources many of which are available from Stock Centers.
- Multinational research community of academic, government and industry laboratories.
Disadvantages
- Does not growth faster as bacteria.
- There are not many parts available for plants in the registry.
- Transforming with Agrobacterium tumefaciens T-DNA integrates less randomly into the plant genome.
- High probabilities of pollution in the medium.
How to use Arabidopsis thaliana?
Successful germination and plant growth requires appropriate soil moisture, nutrient levels, light intensity, humidity, and temperature. If any of these are compromised, A. thaliana will respond by flowering early and dying prematurely and producing little leaf mass for experiments. The plants can also be stressed by overcrowding, fungus infestation, or insect infestation. All these factors must be conscientiously monitored to ensure reliable data obtained from experiments using these plants. Maintenance of soil moisture is imperative for successful seed germination.
The optimal temperature for plant growth is 25°C, with lower temperatures being allowable; higher temperatures can be very detrimental, particularly in the first two weeks of growth.
Growth conditions
With the purpose to obtain several perfects A. thaliana, the Arabidopsis Biological Resource Center (ABCR) has led methods to cultivate and grow these plants inside and outside the laboratory, in medium and soil. These methods and required conditions can be checked in the protocol “Handling Arabidopsis plants and seeds”. The growth and develop of A. thaliana depends of several environmental conditions, besides to the genetic background; with the correct conditions,light, temperature and watering, plants produce flowers within 4-5 weeks and seeds can be harvested 8 to 10 weeks after planting.
With respect to the light, the optimum intensity is 120-150 umol/m2sec; higher intensities may produce death of the seedlings, while low light intensities may produce weak plants without enough chlorophyta. Besides, under photoperiods greater than 12 hours plants flower rapidly.
According to the temperature, 22-23°C ir the optimum condition, lower temperatures can be accepted but higher are impossible. The consequences with higher temperatures may result with the reduce number of the leaves, flowers and seeds while in lower temperatures growth is slow.
Other condition is the water required, the optimal humidity is the mild (50% to 60%). Lower humidity cause drying soil and higher humidity cause plant sterility.
For transformation we used the floral dip method described in this protocol: (Sushanta, 2013).
- 1. Preparation of Agrobacterium strain containing the gene. Inoculate in to a 5 ml LB medium, incubate at 28°C for 2 days.
- 2. Inoculation of culture in a 500 ml LB medium containing antibiotics.
- 3. Centrifugate at 4000rpm for 10 min. at room temperature.
- 4. Resuspended in 1 volume of 5% sucrose solution. 0.02% silwet L-77 is use in the mixing solution and Agrobacterium cell suspension transfer to a 500 ml beaker. Agrobacterium suspension quantity up to 400-500 ml required for transformation of at least six pots of A. thaliana.
- 5. Arabidopsis thaliana plants are invert and dip into the Agrobacterium suspension for 10 seconds with gentle agitation.
- 6. The plants are removed from the solution and wash.
- 7. Plants are wrapping with plastic cover, are laydown on their sides for 16-24 hours to maintain high humidity.
- 8. On the next day, remove the cover and keep it in a growth chamber.
- 9. Plant dry seeds collect using a sieve mesh.
Transformation
The transformation of the plant was made using V9(BBa_k382002) without the construct, this was made by using the floral deeping protocol, this was using an agrobacterium culture transformed with the vector, the flowers of the plant were placed in the medium with agrobacterium and then let it rest.
The plant were placed in a room at 21 degrees but we werent able to prove if the vector was being expressed or not.
Modeling
In 2010 Harvard's 2010 iGarden used its V10 for Agrobacterium transformation and confirmed its presence with kanamycin. This team showed their results for V9 (BBa_K382002) and V10 (BBa_K382003). In their results the vectors were transformed into Arabidopsis, after Agrobacterium mediated transformation, and confirmed their presence with kanamycin.
Because plants take a long time to grow, the function of the parts were unverifiable in Arabidopsis. However E. Coli was used to confirm the transcription and translation of the proteins of interest.
(A) through (D) are normalized plots of miraculin and brazzein YFP-fused constructs expressed in E. Coli. 2xYFP tags were attached to either the N- or C- terminus to ensure that folding was not hindered. In all cases relative YFP fluorescence had appreciably increased after 120 minutes as compared to the non-induced E. Coli
References
- Highered.mheducation.com, (2014). Genetic Portrait Chapters A-E. [online] Available at: http://highered.mheducation.com/sites/007352526x/student_view0/genetic_portrait_chapters_a-e.html [Accessed 26 Sep. 2014].
- Boyle et al.: A BrioBrick compatible strategy for genetic modification of plants. Journal of Biological Engineering 2012 6:8.
- Sushanta, K. (2013). Floral dip: A simple and efficient agrobacterium mediated transformation method is used in a model plant Arabidopsis Thaliana. Cibtech Journal of Bio-Protocols, 14-20.
- Arabidopsis Biological Resource Center, (2014). Handling Arabidopsis plants and seeds. [online] Available at: https://abrc.osu.edu/sites/abrc.osu.edu/files/abrc_handling_seed_2013.pdf [Accessed 27 Sep. 2014].
- Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. (n.d.). Proquest. Retrieved October 12, 2014, from http://0-search.proquest.com.millenium.itesm.mx/docview/204469095/43FC9248199F47A8PQ/11?accountid=11643
 "
"