Team:Paris Bettencourt/Project/Odor Library
From 2014.igem.org
| (172 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | |||
{{Team:Paris_Bettencourt/Menu}} | {{Team:Paris_Bettencourt/Menu}} | ||
| Line 11: | Line 10: | ||
background : rgb(255,0,99); | background : rgb(255,0,99); | ||
} | } | ||
| + | |||
#topheader { | #topheader { | ||
width : 100%; | width : 100%; | ||
| Line 28: | Line 28: | ||
h6 { | h6 { | ||
font-family : Arial; | font-family : Arial; | ||
| - | font-size : | + | font-size : 22px; |
font-weight : bold; | font-weight : bold; | ||
| - | margin-left : | + | margin-left : 5%; |
} | } | ||
#logoblanc { | #logoblanc { | ||
| Line 43: | Line 43: | ||
align : center; | align : center; | ||
z-index : 50; | z-index : 50; | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
display : block; | display : block; | ||
} | } | ||
p { | p { | ||
text-align : justify; | text-align : justify; | ||
| - | |||
background-color : transparent; | background-color : transparent; | ||
} | } | ||
#headtable { | #headtable { | ||
| - | width : | + | width : 92.5%; |
| - | height : | + | height : 350px; |
| - | + | background : rgb(255,0,99); | |
| - | background : rgb( | + | margin-right : 3.75%; |
| + | margin-left : 3.75%; | ||
} | } | ||
#headtable tr td { | #headtable tr td { | ||
| - | width : | + | width : 33.333%; |
heigth : 100%; | heigth : 100%; | ||
color : rgb(255,255,255); | color : rgb(255,255,255); | ||
| - | font-size : | + | font-size : 14px; |
| + | align : justify; | ||
} | } | ||
| + | #headtable tr td a { | ||
| + | color : rgb(193,192,197); | ||
| + | } | ||
#tablelien { | #tablelien { | ||
width : 100%; | width : 100%; | ||
| - | height : | + | height : 80px; |
text-align : center; | text-align : center; | ||
} | } | ||
| Line 92: | Line 77: | ||
font-size : 16px; | font-size : 16px; | ||
} | } | ||
| - | # | + | #tablelien tr td a { |
| + | color : rgb(197,192,193); | ||
| + | } | ||
| + | #fond { | ||
| + | width : 100% | ||
| + | height : 350px; | ||
| + | background : rgb(255,0,99); | ||
| + | } | ||
| + | .project { | ||
width : 100%; | width : 100%; | ||
| - | |||
display : inline-block; | display : inline-block; | ||
} | } | ||
| - | + | .project .text1 { | |
display : inline-block; | display : inline-block; | ||
vertical-align : middle; | vertical-align : middle; | ||
position : relative; | position : relative; | ||
| - | width : | + | width : 43%; |
| - | margin-left : | + | margin-left : 4%; |
| - | font-size : | + | font-size : 15px; |
| + | float : left; | ||
} | } | ||
| - | + | .project .text2 { | |
| - | + | float : right; | |
| - | width : | + | width : 43%; |
| - | + | margin-right : 4%; | |
| - | + | font-size : 15px; | |
| - | + | ||
| - | + | ||
} | } | ||
| - | + | .project .text2 img { | |
| + | width : 100%; | ||
| + | } | ||
| + | .project .text1 img { | ||
| + | width : 100%; | ||
| + | } | ||
| + | #reference { | ||
width : 100%; | width : 100%; | ||
| - | height : | + | height : 350px; |
display : inline-block; | display : inline-block; | ||
} | } | ||
| - | # | + | #reference .text1 { |
| + | position : absolute; | ||
display : inline-block; | display : inline-block; | ||
vertical-align : middle; | vertical-align : middle; | ||
position : relative; | position : relative; | ||
| - | width : | + | width : 44%; |
| - | margin-left : | + | margin-left : 5%; |
| + | font-size : 12px; | ||
} | } | ||
| - | # | + | #headtable tr td .text1 ul { |
| + | text-align : left; | ||
| + | } | ||
| + | #reference .text2 { | ||
position : absolute; | position : absolute; | ||
| - | width : | + | width : 44%; |
| - | height : | + | height : 250px; |
| - | + | left : 51%; | |
| - | + | margin-top : 60px; | |
| - | + | font-size : 12px; | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
} | } | ||
#top { | #top { | ||
position : fixed; | position : fixed; | ||
| - | right : | + | right : 20px; |
bottom : 10px; | bottom : 10px; | ||
| + | width : 25px; | ||
| + | height : 25px; | ||
z-index : 100; | z-index : 100; | ||
} | } | ||
| + | #top img { | ||
| + | width : 100%; | ||
| + | height : 100%; | ||
| + | } | ||
.nameimg { | .nameimg { | ||
position : absolute; | position : absolute; | ||
| - | width : | + | width : 300px; |
height : 200px; | height : 200px; | ||
margin-left : 10%; | margin-left : 10%; | ||
| Line 192: | Line 160: | ||
margin-right : 5%; | margin-right : 5%; | ||
} | } | ||
| + | #headtable tr td b { | ||
| + | font-size : 18px; | ||
| + | } | ||
| + | .playlist { | ||
| + | position : absolute; | ||
| + | color : rgb(255,0,99); | ||
| + | top : 270px; | ||
| + | left : 5%; | ||
| + | } | ||
| + | .playlist img { | ||
| + | width : 20px; | ||
| + | height : 20px; | ||
| + | } | ||
| + | .lower { | ||
| + | font-size : 12px; | ||
| + | } | ||
| + | #fond ul { | ||
| + | list-style-image : url("https://static.igem.org/mediawiki/2014/f/f2/MicodonPB.png"); | ||
| + | } | ||
| + | #link { | ||
| + | color : rgb(255,0,99); | ||
| + | } | ||
| + | #alignliste { | ||
| + | text-align : justify; | ||
| + | } | ||
| + | table.tableizer-table { | ||
| + | border: 1px solid #CCC; font-family: Arial, Helvetica, sans-serif | ||
| + | font-size: 12px; | ||
| + | } | ||
| + | .tableizer-table td { | ||
| + | padding: 4px; | ||
| + | margin: 3px; | ||
| + | border: 1px solid #ccc; | ||
| + | } | ||
| + | .tableizer-table th { | ||
| + | background-color: #FA0063; | ||
| + | color: #FFF; | ||
| + | font-weight: bold; | ||
| + | } | ||
</style> | </style> | ||
<body> | <body> | ||
| - | <p id=top><a href="#topheader"> | + | <p id=top><a href="#topheader"><img src="https://static.igem.org/mediawiki/2014/4/41/Arrow.png"></a></p> |
| - | <div id="topheader"> | + | <div id="topheader"><a id=link class=playlist href="https://www.youtube.com/watch?v=h3a5vN4tUl4" target="_blank"><img src="https://static.igem.org/mediawiki/2014/0/05/Playlistpb.png"> Stop And Smell The Roses</a> |
<img src="https://static.igem.org/mediawiki/2014/b/b1/Smelltheroses.png" class=nameimg> | <img src="https://static.igem.org/mediawiki/2014/b/b1/Smelltheroses.png" class=nameimg> | ||
</div> | </div> | ||
| - | <div id=logo><img id=logoblanc src="https://static.igem.org/mediawiki/2014/7/7d/Logoblanc.png">< | + | <div id=logo><img id=logoblanc src="https://static.igem.org/mediawiki/2014/7/7d/Logoblanc.png"></div> |
| - | + | <div id=fond> | |
| + | <table id=headtable> | ||
<tr> | <tr> | ||
| - | <td><b> | + | <td><b><center>BACKGROUND</center></b></br><br> |
| - | <p class=text1> | + | <p class=text1>Synthetic enzymes can produce odors that humans experience directly, without special instruments. The banana and wintergreeen smell BioBricks are iGEM icons, and a favorite way to introduce genetic engineering. An expanded library of easy-to-use odor enzymes would take synthetic biology to new audiences for creativity, beauty and fun!</p></td> |
| - | <td><b> | + | <td><b><center>AIMS</center></b></br><br> |
| - | <p class=text1> | + | <p class=text1><left>We standardized and simplified existing smell-producing BioBricks for banana, wintergreen, lemon and rain. Also, we created new BioBricks for the aromas of popcorn and jasmine. We made an odor wheel made out of genetic odors that follow a standard organization so that the general public can play with odor genes.</left></li></ul></p></td> |
| - | <td><b> | + | <td><b><center>ACHIEVEMENTS</center></b></br><br> |
| - | <p class=text1> | + | <p class=text1><ul><li> Created BioBricks coding for sequences for different enzymes to nullify the bad odor produced by <i>E. coli.</i></li> <li>We produced the smells that compose the main odor categories perceived by humans.</li> <li>BioBricks submitted to be BioBrick registry: <a href="http://parts.igem.org/Part:BBa_K1403003">BBa_K1403003</a>, <a href="http://parts.igem.org/Part:BBa_K1403006">BBa_K1403006</a>, <a href="http://parts.igem.org/Part:BBa_K1403009">BBa_K1403009</a>, <a href="http://parts.igem.org/Part:BBa_K1403012">BBa_K1403012</a>, <a href="http://parts.igem.org/Part:BBa_K1403017">BBa_K1403017</a>, <a href="http://parts.igem.org/Part:BBa_K1403019">BBa_K1403019</a></li> </p></td> |
| - | + | ||
| - | < | + | |
</tr> | </tr> | ||
</table> | </table> | ||
| + | </div> | ||
<table id=tablelien> | <table id=tablelien> | ||
<tr> | <tr> | ||
| - | <td><a href="# | + | |
| - | <td><a href="# | + | <td><a href="#part1">Introduction</a></td> |
| - | + | <td><a href="#part2">Results</a></td> | |
| - | + | <td><a href="#part3">Methods</a></td> | |
| - | <td><a href="# | + | |
</tr> | </tr> | ||
</table> | </table> | ||
| - | <div id= | + | <div id=part1 class=project> |
| + | |||
| + | <p class=text2> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/5/56/ODOR_WHEEL.png"> | ||
| + | <b>Figure 1. Odor palette composed of BioBricks containing coding sequences for different enzymes known to catalyze reactions that yield volatile compounds with distinctive smells. We included odors with different tonalities in order to explore the aromas resulting from different combinations of smelly units. The tonality of each smell is categorized as butter, balminess, citrus, non-citrus fruit and herbal. These cover half of the main odor categories perceivable to human beings.</b> </br> | ||
| + | </br></br></br></br></br></br> | ||
| + | </p> | ||
<h6>Introduction</h6> | <h6>Introduction</h6> | ||
| - | <p> | + | <br> |
| - | + | <p class=text1>All living things can detect chemical signals from the environment. In humans, the direct chemical sense takes the form of olfaction. Although our sense of smell is less sensitive than that of other mammals, it is still capable of detecting many compounds at concentrations lower than 1 part per million. Odors can trigger emotional responses and memory associations, making them a very direct and visceral way to experience the environment. <br> | |
| - | + | <br> | |
| - | + | ||
| - | + | Bacterial enzymes can produce volatile compounds, including many that are surprising and fun. A generation of iGEMers has had the chance to experience bacteria that smell like <a href="BBa_J45200">banana</a> or <a href="BBa_J45200">mint</a>. The goal of this project is to expand the range of odors that can be created with synthetic enzymes. An easy-to-use, standardized genetic odor library will empower a new generation to play with synthetic biological smells. | |
| - | + | </p></div> | |
| - | + | <div id=part2 class=project> | |
| - | + | ||
| - | + | <p class=text2> | |
| + | <img src="https://static.igem.org/mediawiki/2014/d/dd/Smellybricks.png" | width='100px'> | ||
| + | <b>Figure 2. Basic structure of the odor cassettes: iGEM prefix, <a href="http://parts.igem.org/Part:BBa_J23108">constitutive promoter</a>, synthetic ribosome-binding site, coding sequence with his tag, and iGEM suffix </b> </br> | ||
| + | </p> | ||
<h6>Results</h6> | <h6>Results</h6> | ||
| - | <p> | + | <br> |
| - | + | <p class=text1> | |
| - | + | We created BioBricks coding for sequences of different enzymes to explore each of the odors described in the palette. These sequences are codon-optmized for produced expression in <i>E. coli</i> and produce the smells that compose the main odor categories perceived by humans. </p> | |
| - | + | </div> | |
| - | <h6> | + | <div id=part3 class=project> |
| - | + | <h6>Methods</h6> | |
| - | + | <br> | |
| + | <p class=text1> | ||
| + | Odorless <i>E. coli</i>: | ||
| + | <br> | ||
| + | The tnaA deletion mutant <i>E. coli</i> was taken from location 63:E:9 in the Keio collection. Standard microbiology techniques were used to culture it in liquid and solid media. Subsequently, a single colony was taken to make a batch of Ca2Cl chemically competent cells using. | ||
| + | <br> | ||
| + | <br> | ||
| + | BioBricks: | ||
| + | <br> | ||
| + | The coding sequences for <a href="http://parts.igem.org/Part:BBa_J45119">BMST1</a>,<a href="http://parts.igem.org/Part:BBa_J45199">ATF1</a>, <a href="http://parts.igem.org/Part:BBa_I742111">LIMS1</a> and <a href="http://parts.igem.org/Part:BBa_K221000">GGS</a> were extracted from the Registry. The ATF1 and LIMS1 generator BioBricks were taken from previous iGEM distribution kits and were cloned into the pSB1C3 vector downstream of BBa_J23108 constitutive promoter. The BMST1, JMT, and GDS sequences were codon optimized for expression in <i>E. coli</i>. A synthetic ribosome-binding site that includes XmaI and AgeI flanking restriction sites was designed using the RBS calculator from the Salis’ lab. All BioBricks were assembled using standard molecular biology cloning techniques and 3A iGEM assembly. | ||
| + | <br> | ||
| + | <br> | ||
| + | Characterization: | ||
| + | <br> | ||
| + | Transformed <i>E. coli</i> was grown in M9 minimal media with 2% glucose and complete amino acid mix overnight with the appropriate substrate. | ||
| + | <table class="tableizer-table"> | ||
| + | <tr class="tableizer-firstrow"><th>System</th><th>Enzyme</th><th>Substrate</th><th>Concentration</th><th>Product</th></tr> | ||
| + | <tr><td>Jasmine</td><td>JMT</td><td>Jasmonic acid</td><td>1 mM</td><td>Methyl jasmonate</td></tr> | ||
| + | <tr><td>Mint</td><td>BMST1</td><td>Salicylic acid</td><td>2 mM</td><td>Methyl salicylate</td></tr> | ||
| + | <tr><td>Banana</td><td>ATF1</td><td>Isoamyl alcohol</td><td>5 mM</td><td>Isoamyl acetate</td></tr> | ||
| + | <tr><td>Limonene</td><td>LIMS1</td><td>Farnesyl diphosphate</td><td>Natural</td><td>(+)-Limonene</td></tr> | ||
| + | <tr><td>Rain</td><td>G/G D-synthase</td><td>Farnesyl diphosphate</td><td>Natural</td><td>Geosmin</td></tr> | ||
| + | <tr><td>Flowers</td><td>BMST1</td><td>Benzoic acid</td><td>5 mM</td><td>Methyl benzoate</td></tr> | ||
| + | <tr><td>Butter</td><td>AldB</td><td>Acetolactic acid</td><td>-</td><td>Acetoin</td></tr> | ||
| + | <tr><td>Sweat</td><td>agaA</td><td>3-M-2-hexenoic acid</td><td>-</td><td>3-HO-3-Methylhexanoic acid</td></tr> | ||
| + | </table> | ||
| + | <b>Table 1. Substrate concentrations (mM).</b> </br> | ||
| + | |||
| + | </p> | ||
</div> | </div> | ||
</body> | </body> | ||
</html> | </html> | ||
| + | {{:Team:Paris_Bettencourt/Footer}} | ||
Latest revision as of 15:56, 3 December 2014

Synthetic enzymes can produce odors that humans experience directly, without special instruments. The banana and wintergreeen smell BioBricks are iGEM icons, and a favorite way to introduce genetic engineering. An expanded library of easy-to-use odor enzymes would take synthetic biology to new audiences for creativity, beauty and fun! |
|
| Introduction | Results | Methods |
 Figure 1. Odor palette composed of BioBricks containing coding sequences for different enzymes known to catalyze reactions that yield volatile compounds with distinctive smells. We included odors with different tonalities in order to explore the aromas resulting from different combinations of smelly units. The tonality of each smell is categorized as butter, balminess, citrus, non-citrus fruit and herbal. These cover half of the main odor categories perceivable to human beings.
Figure 1. Odor palette composed of BioBricks containing coding sequences for different enzymes known to catalyze reactions that yield volatile compounds with distinctive smells. We included odors with different tonalities in order to explore the aromas resulting from different combinations of smelly units. The tonality of each smell is categorized as butter, balminess, citrus, non-citrus fruit and herbal. These cover half of the main odor categories perceivable to human beings.
Introduction
All living things can detect chemical signals from the environment. In humans, the direct chemical sense takes the form of olfaction. Although our sense of smell is less sensitive than that of other mammals, it is still capable of detecting many compounds at concentrations lower than 1 part per million. Odors can trigger emotional responses and memory associations, making them a very direct and visceral way to experience the environment.
Bacterial enzymes can produce volatile compounds, including many that are surprising and fun. A generation of iGEMers has had the chance to experience bacteria that smell like banana or mint. The goal of this project is to expand the range of odors that can be created with synthetic enzymes. An easy-to-use, standardized genetic odor library will empower a new generation to play with synthetic biological smells.
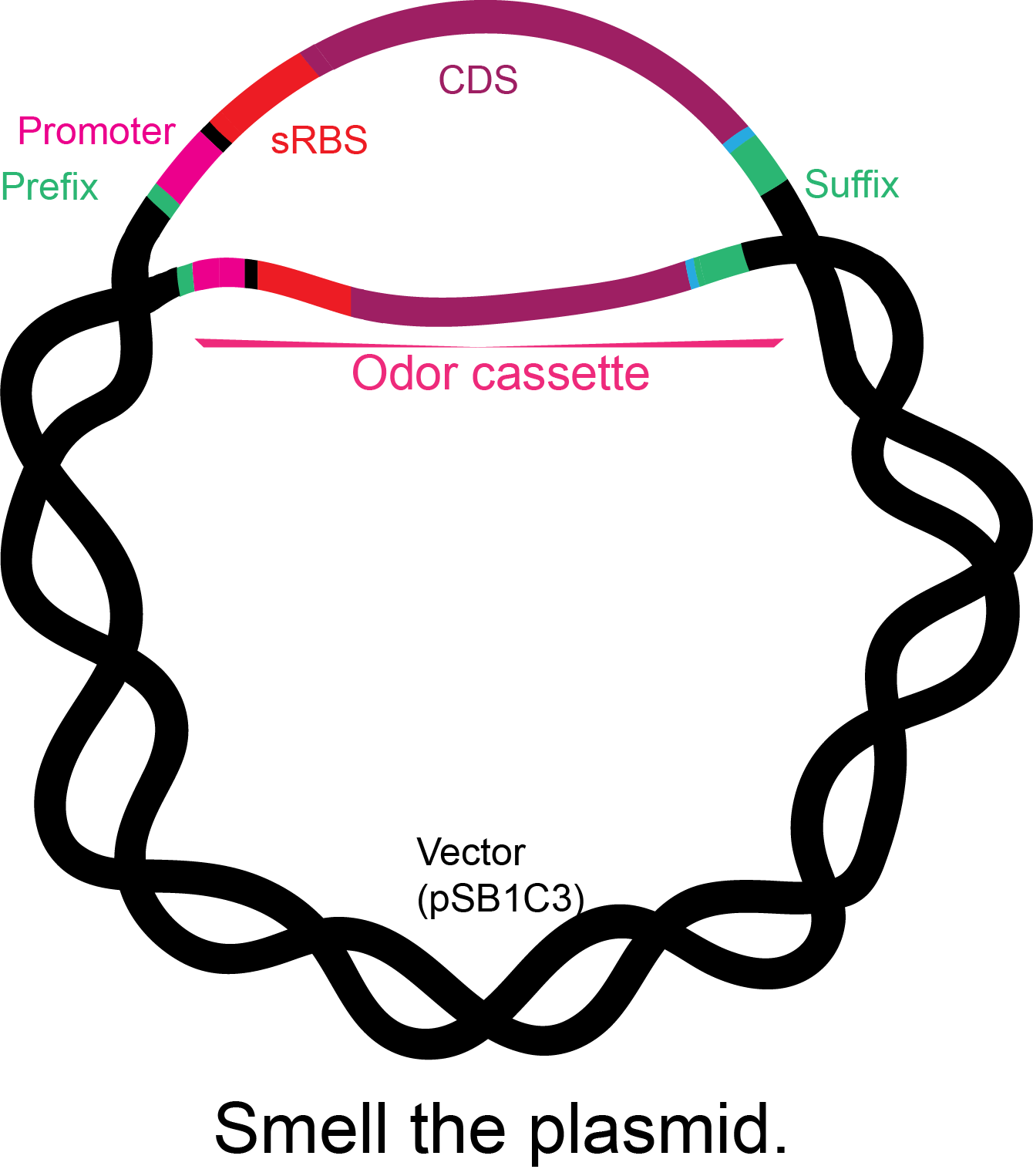 Figure 2. Basic structure of the odor cassettes: iGEM prefix, constitutive promoter, synthetic ribosome-binding site, coding sequence with his tag, and iGEM suffix
Figure 2. Basic structure of the odor cassettes: iGEM prefix, constitutive promoter, synthetic ribosome-binding site, coding sequence with his tag, and iGEM suffix
Results
We created BioBricks coding for sequences of different enzymes to explore each of the odors described in the palette. These sequences are codon-optmized for produced expression in E. coli and produce the smells that compose the main odor categories perceived by humans.
Methods
Odorless E. coli:
The tnaA deletion mutant E. coli was taken from location 63:E:9 in the Keio collection. Standard microbiology techniques were used to culture it in liquid and solid media. Subsequently, a single colony was taken to make a batch of Ca2Cl chemically competent cells using.
BioBricks:
The coding sequences for BMST1,ATF1, LIMS1 and GGS were extracted from the Registry. The ATF1 and LIMS1 generator BioBricks were taken from previous iGEM distribution kits and were cloned into the pSB1C3 vector downstream of BBa_J23108 constitutive promoter. The BMST1, JMT, and GDS sequences were codon optimized for expression in E. coli. A synthetic ribosome-binding site that includes XmaI and AgeI flanking restriction sites was designed using the RBS calculator from the Salis’ lab. All BioBricks were assembled using standard molecular biology cloning techniques and 3A iGEM assembly.
Characterization:
Transformed E. coli was grown in M9 minimal media with 2% glucose and complete amino acid mix overnight with the appropriate substrate.
| System | Enzyme | Substrate | Concentration | Product |
|---|---|---|---|---|
| Jasmine | JMT | Jasmonic acid | 1 mM | Methyl jasmonate |
| Mint | BMST1 | Salicylic acid | 2 mM | Methyl salicylate |
| Banana | ATF1 | Isoamyl alcohol | 5 mM | Isoamyl acetate |
| Limonene | LIMS1 | Farnesyl diphosphate | Natural | (+)-Limonene |
| Rain | G/G D-synthase | Farnesyl diphosphate | Natural | Geosmin |
| Flowers | BMST1 | Benzoic acid | 5 mM | Methyl benzoate |
| Butter | AldB | Acetolactic acid | - | Acetoin |
| Sweat | agaA | 3-M-2-hexenoic acid | - | 3-HO-3-Methylhexanoic acid |
 "
"






 Stop And Smell The Roses
Stop And Smell The Roses

 +33 1 44 41 25 22/25
+33 1 44 41 25 22/25


