Team:AMU-Poznan/Project
From 2014.igem.org
(Difference between revisions)
| (34 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | { | + | <html> |
| + | <style> | ||
| + | body { | ||
| + | background-color: #dddddd; | ||
| + | } | ||
| + | </style> | ||
| + | <head> | ||
| + | <meta http-equiv="Content-Type" content="text/html;charset=UTF-8" > | ||
| + | </head> | ||
| - | < | + | <body> |
| + | <table id="menu" width="100%" cellspacing="0" height="135px"> | ||
| + | <tr> | ||
| + | <td bgColor="#FFFFFF"></td> | ||
| + | <td valign="bottom" width="975px" align="center" bgColor="#FFFFFF"> | ||
| + | <table align="center"> | ||
| + | |||
| + | <tr height="50px"> | ||
| + | <td width="162px" align="center" onMouseOver="this.bgColor='#FEE5AD'" onMouseOut="this.bgColor='#e9eaed'" bgColor=#e9eaed > | ||
| + | <a href="https://2014.igem.org/Team:AMU-Poznan"style="text-decoration:none;color:#1C140D">HOME </a> </td> | ||
| + | |||
| + | <td width="162px" align="center" onMouseOver="this.bgColor='#FEE5AD'" onMouseOut="this.bgColor='#e9eaed'" bgColor=#e9eaed > | ||
| + | <a href="https://2014.igem.org/Team:AMU-Poznan/Project" style="text-decoration:none;color:#1C140D">PROJECT</a> </td> | ||
| + | |||
| + | <td width="162px" align="center" onMouseOver="this.bgColor='#FEE5AD'" onMouseOut="this.bgColor='#e9eaed'" bgColor=#e9eaed > | ||
| + | <a href="https://2014.igem.org/Team:AMU-Poznan/Parts"style="text-decoration:none;color:#1C140D">PARTS</a></td> | ||
| + | |||
| + | <td width="162px" align="center" onMouseOver="this.bgColor='#FEE5AD'" onMouseOut="this.bgColor='#e9eaed'" bgColor=#e9eaed > | ||
| + | <a href="https://2014.igem.org/Team:AMU-Poznan/Team" style="text-decoration:none;color:#1C140D">TEAM </a> </td> | ||
| - | < | + | <td width="162px" align="center" onMouseOver="this.bgColor='#FEE5AD'" onMouseOut="this.bgColor='#e9eaed'" bgColor=#e9eaed > |
| - | + | <a href="https://igem.org/Team.cgi?year=2014&team_name=AMU-Poznan" style="text-decoration:none;color:#1C140D">OFFICIAL TEAM PROFILE</a> </td> | |
| - | < | + | <td width="162px" align="center" onMouseOver="this.bgColor='#FEE5AD'" onMouseOut="this.bgColor='#e9eaed'" bgColor=#e9eaed > |
| + | <a href="https://2014.igem.org/Team:AMU-Poznan/Notebook" style="text-decoration:none;color:#1C140D">NOTEBOOK </a> </td> | ||
| - | + | <td width="162px" align="center" onMouseOver="this.bgColor='#FEE5AD'" onMouseOut="this.bgColor='#e9eaed'" bgColor=#e9eaed > | |
| - | <td align="center" | + | <a href="https://2014.igem.org/wiki/index.php?title=Team:AMU-Poznan/Safety" style="text-decoration:none;color:#1C140D">SAFETY </a></td> |
| - | < | + | <td width="162px" align="center" onMouseOver="this.bgColor='#FEE5AD'" onMouseOut="this.bgColor='#e9eaed'" bgColor=#e9eaed > |
| - | + | <a href="https://2014.igem.org/Team:AMU-Poznan/Modeling" style="text-decoration:none;color:#1C140D">MODELING & SURVEY </a></td> | |
| - | < | + | </tr> |
| - | <tr | + | |
| + | <img src="https://static.igem.org/mediawiki/2014/d/d4/Logoamu.png" width="60%"> | ||
| - | < | + | <tr> <td colspan="5"></td> |
| - | < | + | </tr> |
| - | |||
| - | |||
| - | < | + | </table> |
| - | <a | + | <br> <br> |
| + | <div class="home"> | ||
| + | <h1>sh-miR Designer v2.0</h1> | ||
| + | Sh-miR designer is a project we started during iGEM 2013. This year we would like to continue and expand functionality of the software. sh-miR designer v1.0 (link) is aimed to create sh-miR molecules based on siRNAs provided by the user. In sh-miR designer v2.0 only the mRNA number (from NCBI database), which expression should be decreased can be provided. Moreover, we expanded functionality of the software with off-target validation and check of immune motifs and also extended miRNA-shuttles database. | ||
| + | sh-miR Designer is a software aimed for fast and efficient design of effective RNA interference (RNAi) reagents - sh-miRs, also known as artificial miRNAs. sh-miRs are RNA particles whose structure is based on miRNA precursor pri-miRNA, but sequence interacting with transcript is changed depending on research purpose. Maintenance of structure of pri-miRNA is very important to enable cellular processing and therefore ensure functionality of artificial particles. sh-miRs delivered to cells on genetic vectors - plasmids or viral vectors - enter natural RNAi pathway and silence target mRNA. They can be used in genetic therapies and basic biomedical research. | ||
| + | We will provide two applications to access the software, one which require siRNA sequences and the second which require transcript accession number from NCBI database. Each user will receive an account with login (e-mail) and password where he/she would be able to check history of software usage.</br> | ||
| + | <h1>Biological background</h1> | ||
| + | </br> | ||
| + | RNA interference (RNAi) is a physiological process of posttranscriptional gene expression regulation caused by double stranded RNA molecules. This is one of the most willingly used experimental techniques. It is used in experiments designed to explore gene function, in creation cellular models of diseases or in genetic therapy design. Among RNAi technology reagents are siRNA (short interfering RNA) and vector reagents shRNA (short hairpin RNA) and sh-miR (sh- microRNA). RNAi reagents are integrated into endogenous RNAi pathway on different phases. sh-miR reagents are designed in the way to be similar to endogenous miRNA precursors (pri-miRNA), because it is important feature to be recognized by proteins involved in microRNA (miRNA) biogenesis.</br> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/e/e1/Dicer.png"> | ||
| + | </br> | ||
| - | + | shRNA or sh-miR reagents are introduced into cells with usage of genetic vectors – plasmids or viral vectors to ensure intracellular expression. They have huge therapeutic potential beacuse of stable reagent production in cells what enables one time therapeutic dosage. Usage of synthetic siRNA reagents requires often dosage, what makes therapy more difficult, especially in case of Central Nervous System Diseases.</br> | |
| - | < | + | <img src="https://static.igem.org/mediawiki/2014/a/a8/Jaja.png"> |
| + | </br> | ||
| - | + | It was recently shown that RNAi reagents introduces into cells can trigger unwanted non-specific effects, e.g. can interact with different transcripts besides the desired one (so called off-target effect) or induce immunological effect. Because in processing of sh-miR reagents endogenous endonucleasis Drosha and Dicer are involved, in case of high expression of reagent they can saturate those proteins as well as Exportin-5 transport protein responsible for export of sh-miR from nucleus. The saturation can influence endogenous RNAi mechanism. It was shown that sh-miR reagents influence less endogenous RNAi pathway, because of lower level of produced double-stranded RNA (dsRNA), preserving efficiency of translation silencing. They also show longer effect than different types of RNAi reagents. Because of described features sh-miR reagents seems to be good choice when designing efficient and non-toxic RNAi reagents.</br></br> | |
| - | + | ||
| + | sh-miR Designer software was designed sh-miR Designer is a software aimed for fast and efficient design of effective RNA interference (RNAi) reagents - sh-miRs, also known as artificial miRNAs. sh-miRs are RNA particles whose structure is based on miRNA precursor pri-miRNA, but sequence interacting with transcript is changed depending on research purpose. Until now scientists have to use multiple unconnected software to design single sh-miR molecule. It is time consuming process, what limits number of designed molecules for biological testing. There is no such software, which includes all important features as molecule spatial structure. Maintenance of structure of pri-miRNA is very important to enable cellular processing and therefore ensure functionality of artificial particles. This interdisciplinary project includes computer sciences with biomedical science. sh-miRs delivered to cells on genetic vectors - plasmids or viral vectors - enter natural RNAi pathway and silence target mRNA. They can be used in genetic therapies and basic biomedical research. </br></br> | ||
| - | < | + | </br> |
| - | < | + | </br> |
| + | <h1>Standard Parts</h1> | ||
| + | </br> | ||
| + | We are creating a particle which can be inserted into plasmid built by Standard parts from Registry of Standard Biological Parts. The sh-miR molecules are inserted into plasmid backbone downstream chosen promoter. You can add any regulatory sequence you want or maybe a fragment coding fluorescent protein to see if the transfection was succesfull. The molecules are fully compatibile with Standard Biological Parts - what is important for users from iGEM especially :). | ||
| + | <img src="https://static.igem.org/mediawiki/2014/b/bb/Shmir-STANDARD.png" width="60%"></br> | ||
| + | <img src="http://biosettia.com/images/miRNA/lentiviral-mirna-expression-01.png" width="60%"></br> | ||
| + | picture from http://biosettia.com/ | ||
| + | <h1>User Guide</h1> | ||
| + | <div align="left"> | ||
| + | There are two suplementary versions of our software. First one (based on last year's software sh-miR v1.0) enable to design sh-miR molecules based on siRNA sequence and the second design sh-miR molecules when NCBI transcript number (NM_...) is provided. In the second version the user also can set GC content, maximum off-target and immunostimulatory properties. The user can also set which pri-miRNA should be used to design sh-miR (or all available in our miRNA database).</br> | ||
| - | < | + | Please follow video where you can see how to use web version of the software</br> |
| - | < | + | and short description how to use client version of the software</br> |
| + | Both versions are well documented on github and readthedocs (see below)</br> | ||
| + | <h3>Usage via Web server</h1> | ||
| + | go to our website <a href="https://shmir.wmi.amu.edu.pl">shmir.wmi.amu.edu.pl</a> | ||
| + | <div align = "center"> | ||
| + | <iframe width="420" height="315" | ||
| + | src="https://www.youtube.com/embed/D3DO_5NODQo"> | ||
| + | </iframe> | ||
| + | </div></br></br> | ||
| + | <br/>input form:<br/></br></br> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/2/24/Zrzut_ekranu_2014-10-15_o_18.07.36.png" width="60%"></br></br></br> | ||
| + | <br/>results:<br/></br></br> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/8/8a/Zrzut_ekranu_2014-10-15_o_22.19.56.png" width="60%"></br></br></br> | ||
| + | users' history:<br/></br></br> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/c/c2/Zrzut_ekranu_2014-10-15_o_18.09.10.png" width="60%"><br/></br></br> | ||
| + | <h3>Usage via client</h3> | ||
| + | </br>Fold sequence via mfold:</br> | ||
| - | + | ./shmir_client.py mfold ACTGAUUUGAC</br> | |
| - | < | + | Create sh-miR from one siRNA strand (active) or two siRNa strands separated by space.</br> |
| + | First strand is active, both are in 5-3 orientation</br></br> | ||
| - | + | ./shmir_client.py from_sirna UUUGUAUUCGCCCUAGCGC CGCUAUGGCGAAUACAAACA</br> | |
| - | + | Create sh-miR from transcript name.</br> | |
| + | Optional parameters</br> | ||
| + | --min_gc : Minimal "GC" content in strand -- default: 40</br> | ||
| + | --max_gc : Maximal "GC" content in strand -- default: 60</br> | ||
| + | --max_offtarget : Maximal offtarget in strand -- default: 10</br> | ||
| + | --mirna_name : The name of miRNA backbone to use -- default: "all"</br> | ||
| + | --stymulators : One of ["yes", "no", "no_difference"] -- default: "no_difference"</br> | ||
| + | ./shmir_client.py from_transcript NM_001618.3 --mirna_name miR-30a --min_gc 22</br> | ||
| + | </div> | ||
| + | <h1>Technology</h1> | ||
| + | <div align="left"> | ||
| + | </br>The application was built with Python 2.7. It is divided into 3 parts:</br> | ||
| + | RESTful API - the “heart” of our software - it performs all calculations. Our application is built on CentOS (Linux) operating system. To maintain better compatibility with our server we use Vagrant to create Virtual Machine and Puppet to manage infrastructure of our project. All you need to do is: type into console “vagrant up” and after a while everything is ready to work. This subproject is written with following technologies/frameworks/libraries: Flask, Flask-Cache, SqlAlchemy, PostgreSQL, Celery, Flower, Rabbitmq, Redis, BioPython, mfold (lightweight), uwsgi, sphinx, Python testing tools (mock, nose, unittest).</br> | ||
| + | Console client - simple console application which communicates with API using requests library. It has 3 options: mfold (folding structure), from_sirna (create sh-miR from siRNA) and from_transcript (create sh-miR from transcript name).</br> | ||
| + | Website - application written in Django framework. It enables you to use the API via graphical interface. You can also create your own account and save the history of your requests for later usage. | ||
| + | </div> | ||
| + | </br> | ||
| + | <h1>API documentation</h1> | ||
| + | </br>Here you can find documentation from code:</br> | ||
| - | + | API: <a href="https://shmir-api.rtfd.org">shmir-api.rtfd.org</a></br> | |
| - | </ | + | Client: <a href="https://shmir-client.rtfd.org">shmir-client.rtfd.org</a></br> |
| - | </ | + | Webpage: <a href="https://shmir-designer.rtfd.org">shmir-designer.rtfd.org</a></br> |
| - | |||
| - | </ | + | <h1>API for developers</h1> |
| - | </ | + | </br> |
| - | </ | + | Here you can find api for developers: <a href="https://github.com/Nozdi/shmir">https://github.com/Nozdi/shmir</a></br> |
| + | <h1>Releases (re-use of 2013iGEM software)</h1> | ||
| + | <div align="left"> | ||
| + | <h3></br>sh-miR designer v1.0 </br></h3> | ||
| + | First version of the program presented on iGEM 2013 by our team. The user have to provide siRNA sequence (one or both strands). The miRNA database includes 5 miRNAs. </br> | ||
| + | <h3>sh-miR designer v1.1 </br></h3> | ||
| + | Strand discrimination functionality was added based on miRbase. </br> | ||
| + | extended miRNA database </br> | ||
| + | <h3>sh-miR designer v2.0 </br></h3> | ||
| + | siRNA prediction algorithm was added to functionality (the input is NCBI transcript number) </br> | ||
| + | user can choose GC content of siRNA molecule, maximal off-target, if he/she wants immunostimulatory sequences inside the molecule and to choose if he/she wants to include all or chosen scaffold </br> | ||
| + | extended miRNA database</br> | ||
| + | |||
| + | What we are working on: </br> | ||
| + | strand discrimination based on therodynamics of siRNA ends; </br> | ||
| + | preparing construct with flanking restriction enzymes </br> | ||
| + | providing plasmids with parts from standard parts </br> | ||
| + | </div> | ||
| + | <h1>Unit Tests</h1> | ||
| + | </br> | ||
| + | <div align="left"> | ||
| - | < | + | In our application we are using unit tests to test alghoritms, functions and database.</br> |
| - | < | + | We are testing inputs and outputs:</br> |
| - | < | + | <pre> |
| + | def test_validate_gc_content(self):</br> | ||
| + | is_in_range = validate_gc_content('ACGT', 40, 60)</br> | ||
| + | self.assertTrue(is_in_range)</br> | ||
| + | Caught errors:</br> | ||
| + | def multiple_input(self, data, error):</br> | ||
| + | for strand in data:</br> | ||
| + | with self.assertRaises(errors.InputException) as err:</br> | ||
| + | validators.check_input(strand)</br> | ||
| + | self.assertEqual(error, str(err.exception))</br> | ||
| + | Database models:</br> | ||
| + | def test_get_and_put_to_db(self):</br> | ||
| + | new_obj = self.TestModel(id=153)</br> | ||
| + | self.put_to_db(new_obj)</br> | ||
| + | objs = self.get_all(self.TestModel)</br> | ||
| + | self.assertEqual(len(objs), 1)</br> | ||
| + | </br> | ||
| + | </pre> | ||
| + | We are using Mock library to test our functions which are using external API’s to create fake response from server and return them into our function:</br> | ||
| + | <pre> | ||
| + | @patch.object(ncbi_api.Entrez, 'esearch')</br> | ||
| + | @patch.object(ncbi_api.Entrez, 'efetch')</br> | ||
| + | def test_get_data(self, mock_efetch, mock_esearch):</br> | ||
| + | ids = [69]</br> | ||
| + | mock_read = Mock()</br> | ||
| + | mock_read.read.return_value = json.dumps({'esearchresult': {'idlist': ids}})</br> | ||
| + | mock_esearch.return_value = mock_read</br> | ||
| + | ncbi_api.get_data(self.GOOD_MRNA)</br> | ||
| + | mock_esearch.assert_called_with(</br> | ||
| + | term='NM_001128164.1', db=self.DATA_BASE, retmode='json'</br> | ||
| + | )</br> | ||
| + | mock_efetch.assert_called_once_with(</br> | ||
| + | db=self.DATA_BASE, id=ids, rettype='fasta', retmode='text'</br> | ||
| + | )</br> | ||
| + | </br> | ||
| + | </pre> | ||
| + | </div> | ||
| + | <h1>Validation</h1> | ||
| + | <h3>old version shmiR v1.0 (average time)</h3> | ||
| + | </br>real 0m6.854s</br> | ||
| + | user 0m0.063s</br> | ||
| + | sys 0m0.027s</br> | ||
| + | <h3>new version shmiR v1.1 (average times)</h3> | ||
| + | real 0m3.271s</br> | ||
| + | user 0m0.155s</br> | ||
| + | sys 0m0.041s</br> | ||
| + | </br> | ||
| + | </br> | ||
| + | <h3>Designed sh-miR</h3> | ||
| + | </br></br> | ||
| + | for $ ./shmir_client.py from_transcript NM_000546.5 | ||
| + | </br> | ||
| + | 1: backbone: miR-31, score: 106, sequence: ACATGACGGAGGTTGTGAG</br> | ||
| + | pdf: ./results/transcript/e9e8b9a2-7e3b-4b24-b26a-633332a3f6b2/miR-31/7270b2d2-0aca-472b-853e-5a9631164d46</br> | ||
| + | result: </br>CATAACAACGAAGAGGGATGGTATTGCTCCTGTAACTCGGAACTGGAGAGGACATGACGGAGGTTGTGAGCGTTGAACTGGGAACCCTCACAACCTCCGTCA | ||
| + | </br>TGTCTCTCCAGTTCCGAGTTACAGGAGCAATACCAT</br>CCCTCTTCGTTGTTATGCTTTCCTGTCTGACAGCAGCTTGGCTACCTCCGTCCTGTTCCTCCTTGTCTT | ||
| + | </br></br> | ||
| + | 2: backbone: miR-31, score: 106, sequence: ATGTTTTGTAGAGATGGGG</br> | ||
| + | pdf: ./results/transcript/e9e8b9a2-7e3b-4b24-b26a-633332a3f6b2/miR-31/56232916-2649-4eb4-9970-c0b0a08ce64e</br> | ||
| + | result: </br>CATAACAACGAAGAGGGATGGTATTGCTCCTGTAACTCGGAACTGGAGAGGATGTTTTGTAGAGATGGGGCGTTGAACTGGGAACCCCCCATCTCTACA | ||
| + | </br>AAACATCTCTCCAGTTCCGAGTTACAGGAGCAATACCAT</br>CCCTCTTCGTTGTTATGCTTTCCTGTCTGACAGCAGCTTGGCTACCTCCGTCCTGTTCCTCCTTGTCTT | ||
| + | </br></br> | ||
| + | 3: backbone: miR-31, score: 106, sequence: ATTACATGGGGTCTAGAAC</br> | ||
| + | pdf: ./results/transcript/e9e8b9a2-7e3b-4b24-b26a-633332a3f6b2/miR-31/37cd84aa-362c-484d-8681-929043cc9c61</br> | ||
| + | result: </br>CATAACAACGAAGAGGGATGGTATTGCTCCTGTAACTCGGAACTGGAGAGGATTACATGGGGTCTAGAACCGTTGAACTGGGAACCGTTCTAG | ||
| + | </br>ACCCCATGTAATCTCTCCAGTTCCGAGTTACAGGAGCAATACCAT</br>CCCTCTTCGTTGTTATGCTTTCCTGTCTGACAGCAGCTTGGCTACCTCCGTCCTGTTCCTCCTTGTCTT | ||
| + | </br></br> | ||
| + | 4: backbone: miR-31, score: 106, sequence: ACCATCATCACACTGGAAG</br> | ||
| + | pdf: ./results/transcript/e9e8b9a2-7e3b-4b24-b26a-633332a3f6b2/miR-31/50b763f6-2193-4743-96c0-61601880e574</br> | ||
| + | result: </br>CATAACAACGAAGAGGGATGGTATTGCTCCTGTAACTCGGAACTGGAGAGGACCATCATCACACTGGAAGCGTTGAACTGGGAACCCTT | ||
| + | </br>CCAGTGTGATGATGGTCTCTCCAGTTCCGAGTTACAGGAGCAATACCAT</br>CCCTCTTCGTTGTTATGCTTTCCTGTCTGACAGCAGCTTGGCTACCTCCGTCCTGTTCCTCCTTGTCTT | ||
| + | </br></br> | ||
| + | 5: backbone: miR-31, score: 106, sequence: ATCTCACCCCATCCCACAC</br> | ||
| + | pdf: ./results/transcript/e9e8b9a2-7e3b-4b24-b26a-633332a3f6b2/miR-31/019ce915-0282-4d83-9538-93c36c790fa8</br> | ||
| + | result: </br>CATAACAACGAAGAGGGATGGTATTGCTCCTGTAACTCGGAACTGGAGAGGATCTCACCCCATCCCACACCGTTGAAC | ||
| + | </br>TGGGAACCGTGTGGGATGGGGTGAGATCTCTCCAGTTCCGAGTTACAGGAGCAATACCAT</br>CCCTCTTCGTTGTTATGCTTTCCTGTCTGACAGCAGCTTGGCTACCTCCGTCCTGTTCCTCCTTGTCTT</br> | ||
| + | </br> | ||
| + | see example pdf with structure <a href="https://static.igem.org/mediawiki/2014/d/da/Sequence_1.pdf">link</a> | ||
| + | </br> | ||
| + | During sh-miR designer development, colleagues from befriended lab used our software, so we had constant feedback from potential users,</br> | ||
| + | who by the way, have nothing to do with bioinformatics. So it's really hard to satisfy their strong need for easy interface.</br> | ||
| + | We also had significant feedback from our survey (see Modeling tab), so we knew what we should emphasise on during develoment.</br> | ||
| + | </br> | ||
| + | <h1>Test cases</h1> | ||
| + | </br>1) ATXN3 - therapeutic sh-miR against SCA3 disease</br> | ||
| + | transcript number: NM_001164782.1 </br> | ||
| + | GC content: default </br> | ||
| + | Off-target: 10 </br> | ||
| + | scaffold: ALL </br> | ||
| + | immunostimulatory: NO</br> | ||
| + | 2) P54 - therapeutic sh-miR against cancer</br> | ||
| + | transcript number: NM_000546.5 </br> | ||
| + | GC content: default </br> | ||
| + | Off-target: 10 </br> | ||
| + | scaffold: ALL </br> | ||
| + | immunostimulatory: YES</br> | ||
| + | 3) biological function analysis, PARP1, we have miR-30a in our lab</br> | ||
| + | transcript number: NM_001618.3 </br> | ||
| + | GC content: default </br> | ||
| + | Off-target: 0 </br> | ||
| + | scaffold: miR-30a </br> | ||
| + | immunostimulatory: no_difference (default) </br> | ||
| + | 4) biological function, PARP1, we have miR-30a in our laboratory and have immune cells</br> | ||
| + | transcript number: NM_001618.3 </br> | ||
| + | GC content: default </br> | ||
| + | Off-target: 0 </br> | ||
| + | scaffold: miR-30a </br> | ||
| + | immunostimulatory: NO</br> | ||
| + | 5) biological function, PARP1 and have immune cells</br> | ||
| + | transcript number: NM_001618.3 </br> | ||
| + | GC content: default </br> | ||
| + | Off-target: 0 </br> | ||
| + | scaffold: ALL </br> | ||
| + | immunostimulatory: NO</br> | ||
| + | <h1>References</h1> | ||
| + | </br> | ||
| + | <div align="left"> | ||
| + | 1. Boudreau RL, Monteys AM, Davidson BL. Minimizing variables among hairpin-based RNAi | ||
| + | vectors reveals the potency of shRNAs. RNA. 2008 Sep;14(9):1834-44. Epub 2008 Aug 12.</br> | ||
| + | 2. Chang K, Elledge SJ, Hannon GJ. Lessons from Nature: microRNA-based shRNA libraries. Nat | ||
| + | Methods. 2006 Sep;3(9):707-14.</br> | ||
| + | 3. Dow LE, Premsrirut PK, Zuber J, Fellmann C, McJunkin K, Miething C, Park Y, Dickins RA, | ||
| + | Hannon GJ, Lowe SW. A pipeline for the generation of shRNA transgenic mice. Nat Protoc. | ||
| + | 2012 Feb 2;7(2):374-93. doi: 10.1038/nprot.2011.446.</br> | ||
| + | 4. Fahim M, Larkin PJ. Designing Effective amiRNA and Multimeric amiRNA Against Plant | ||
| + | Viruses. Methods Mol Biol. 2013;942:357-77. doi: 10.1007/978-1-62703-119-6_19.</br> | ||
| + | 5. Fan ZD, Zhang L, Shi Z, Gan XB, Gao XY, Zhu GQ. Artificial microRNA interference targeting | ||
| + | AT(1a) receptors in paraventricular nucleus attenuates hypertension in rats. Gene Ther. 2012 | ||
| + | Aug;19(8):810-7. doi: 10.1038/gt.2011.145. Epub 2011 Sep 29.</br> | ||
| + | 6. Fellmann C, Zuber J, McJunkin K, Chang K, Malone CD, Dickins RA, Xu Q, Hengartner MO, | ||
| + | Elledge SJ, Hannon GJ, Lowe SW. Functional identification of optimized RNAi triggers using a | ||
| + | massively parallel sensor assay. Mol Cell. 2011 Mar 18;41(6):733-46. Epub 2011 Feb 25.</br> | ||
| + | 7. Maczuga P, Lubelski J, van Logtenstein R, Borel F, Blits B, Fakkert E, Costessi A, Butler D, van | ||
| + | Deventer S, Petry H, Koornneef A, Konstantinova P. Embedding siRNA sequences targeting | ||
| + | Apolipoprotein B100 in shRNA and miRNA scaffolds results in differential processing and in | ||
| + | vivo efficacy. Mol Ther. 2012 Oct 23. doi: 10.1038/mt.2012.160. [Epub ahead of print]</br> | ||
| + | 8. Maczuga P, Koornneef A, Borel F, Petry H, van Deventer S, Ritsema T, Konstantinova P. | ||
| + | Optimization and comparison of knockdown efficacy between polymerase II expressed | ||
| + | shRNA and artificial miRNA targeting luciferase and Apolipoprotein B100. BMC | ||
| + | Biotechnol. 2012 Jul 24;12:42. doi: 10.1186/1472-6750-12-42.</br> | ||
| + | 9. Matveeva OV, Nazipova NN, Ogurtsov AY, Shabalina SA. Optimized models for design of | ||
| + | efficient miR30-based shRNAs. Front Genet. 2012;3:163. Epub 2012 Aug 29.</br> | ||
| + | 10. McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, Gilmore BL, Burstein | ||
| + | H, Peluso RW, Polisky B, Carter BJ, Davidson BL. Artificial miRNAs mitigate shRNA-mediated | ||
| + | toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci | ||
| + | U S A. 2008 Apr 15;105(15):5868-73. Epub 2008 Apr 8.</br> | ||
| + | 11. Schwab R, Ossowski S, Warthmann N, Weigel D. Directed gene silencing with artificial | ||
| + | microRNAs. Methods Mol Biol. 2010;592:71-88.</br> | ||
| + | 12. Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach | ||
| + | MR, Sheth N, Bradshaw J, Burchard J, Kulkarni A, Cavet G,Sachidanandam R, McCombie WR, Cleary MA, Elledge SJ, Hannon GJ. Second-generation shRNA libraries | ||
| + | covering the mouse and human genomes. Nat Genet. 2005 Nov;37(11):1281-8. Epub 2005 | ||
| + | Oct 2.</br> | ||
| + | 13. Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for | ||
| + | single-copy polymerase II-regulated RNA interference in mammalian cells. Proc Natl Acad Sci | ||
| + | U S A. 2005 Sep 13;102(37):13212-7. Epub 2005 Sep 1.</br> | ||
| + | 14. Wang X, Yang Y, Zhou J, Yu C, Cheng Y, Yan C, Chen J.Two-step method for constructing | ||
| + | Arabidopsis artificial microRNA vectors. Biotechnol Lett. 2012 Jul;34(7):1343-9. Epub 2012 | ||
| + | Mar 22.</br> | ||
| + | 15. Warthmann N, Ossowski S, Schwab R, Weigel D. Artificial MicroRNAs for Specific Gene | ||
| + | Silencing in Rice. Methods Mol Biol. 2013;956:131-49. doi: 10.1007/978-1-62703-194-3_11.</br> | ||
| + | 16. Zeng Y, Cullen BR. Efficient processing of primary microRNA hairpins by Drosha requires | ||
| + | flanking nonstructured RNA sequences. J Biol Chem. 2005 Jul 29;280(30):27595-603. Epub | ||
| + | 2005 Jun 1.</br> | ||
| + | 17. Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the | ||
| + | expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002 Jun;9(6):1327- | ||
| + | 33</br> | ||
| + | </div> | ||
| + | <h1>Judging Form (in <b>bold</b> what we did to achieve gold medal) </h1> | ||
| + | </br> | ||
| + | <div align="left">Bronze. The following 4 goals must be achieved:</br> | ||
| + | <ul> | ||
| + | <li><b>Register the team, have a great summer, and have fun attending the Jamboree in Boston.</br></li> | ||
| + | <li>Create and share a description of the team's project via the iGEM wiki.</br></b></li> | ||
| + | <li>Present a Poster and Talk at the Regional Jamboree and World Championship Jamboree.<b> We will on Jamboree :)</br></li> | ||
| + | <li>Develop and make available, via The iGEM Software Registry, an open source software that supports Synthetic Biology based on Standard Parts.</li></b></br></br> | ||
| + | </ul> | ||
| + | Silver: In addition to the Bronze Medal requirements, the following 4 goals must be achieved:</br> | ||
| + | <ul> | ||
| + | <b><li>Demonstrate the relevance of your development for Synthetic Biology based on standard Parts.</br></li> | ||
| + | <li>Provide a comprehensive and well-designed User Guide. (Be creative! An instructional video may work as well.)</br></li> | ||
| + | <li>Provide detailed API documentation, preferably, automatically built from source code documentation (use tools like doxygen, yard, rdoc, naturaldocs, etc).</br></li> | ||
| + | <li>Demonstrate that you followed best practises in software development so that other developers can modify, use and reuse your code. Provide more than one realistic test case. Examples of best practices are automated unit testing and documentation of test coverage , bug tracking facilities, documentation of releases and changes between releases.</br></br></b></li> | ||
| + | </ul> | ||
| + | Gold: In addition to the Bronze and Silver Medal requirements, two additional goals must be achieved: </br> | ||
| + | <b><ul><li>Provide a convincing validation, testing the performance of the development -- experimentally (can be outsourced) or by other teams and users. Note, even if the algorithm or tool turns out not to work that well, the Gold requirement is fulfilled if the test is good and the analysis convincing. Validation may include: preferably experiments, informatics analysis (complexity, run time) of an algorithm, or user studies.</li></ul></br></b> | ||
| + | And the second goal can be any one of the following:</br> | ||
| + | <ul> | ||
| + | <li>Make your software interact / interface with the Registry.</br></li> | ||
| + | <li><b>Re-use and further develop previous iGEM software projects (or parts thereof) or use and/or improvement of existing synthetic biology tools or frameworks.</br></li> | ||
| + | <li>Develop a well documented library or API for other developers (rather than “only” a stand-alone app for end users.)</b></br></li> | ||
| + | <li>Support and use the SBOL and / or SBOLv standard.</br></li> | ||
| + | <li><b>iGEM projects involve important questions beyond the bench, for example relating to (but not limited to) ethics, sustainability, social justice, safety, security, or intellectual property rights. Describe an approach that your team used to address at least one of these questions. Evaluate your approach, including whether it allowed you to answer your question(s), how it influenced the team’s scientific project, and how it might be adapted for others to use (within and beyond iGEM). We encourage thoughtful and creative approaches, and those that draw on past Policy & Practice (formerly Human Practices) activities.</li> | ||
| + | </ul></b></br></br> | ||
| + | </div></div> | ||
| + | <div class="footer"> | ||
| + | <br> | ||
| + | <hr> | ||
| + | <a href="https://igem.org/"><img src="https://static.igem.org/mediawiki/igem.org/6/60/Igemlogo_300px.png" width="55px"></a> | ||
| + | <a href="https://github.com/igemsoftware/AMU-Poznan2013"><img src="https://assets-cdn.github.com/images/modules/logos_page/GitHub-Mark.png" width="55px"></a> | ||
| + | <a href="http://international.amu.edu.pl/"><img src="http://siw.amu.edu.pl/__data/assets/file/0005/162752/logo_wersja-uzupeniajca_czarny_2.jpg" width="55px"></a> | ||
| + | <br> | ||
| + | </div> | ||
| + | </td> | ||
| + | <td bgColor="#FFFFFF"></td> | ||
| + | </tr> | ||
| + | </table> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | </table> | |
| - | + | ||
| - | + | ||
| - | </ | + | |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | </ | + | </body> |
| - | |||
| - | |||
| - | |||
</html> | </html> | ||
Latest revision as of 21:18, 17 October 2014
sh-miR Designer v2.0Sh-miR designer is a project we started during iGEM 2013. This year we would like to continue and expand functionality of the software. sh-miR designer v1.0 (link) is aimed to create sh-miR molecules based on siRNAs provided by the user. In sh-miR designer v2.0 only the mRNA number (from NCBI database), which expression should be decreased can be provided. Moreover, we expanded functionality of the software with off-target validation and check of immune motifs and also extended miRNA-shuttles database. sh-miR Designer is a software aimed for fast and efficient design of effective RNA interference (RNAi) reagents - sh-miRs, also known as artificial miRNAs. sh-miRs are RNA particles whose structure is based on miRNA precursor pri-miRNA, but sequence interacting with transcript is changed depending on research purpose. Maintenance of structure of pri-miRNA is very important to enable cellular processing and therefore ensure functionality of artificial particles. sh-miRs delivered to cells on genetic vectors - plasmids or viral vectors - enter natural RNAi pathway and silence target mRNA. They can be used in genetic therapies and basic biomedical research. We will provide two applications to access the software, one which require siRNA sequences and the second which require transcript accession number from NCBI database. Each user will receive an account with login (e-mail) and password where he/she would be able to check history of software usage.Biological backgroundRNA interference (RNAi) is a physiological process of posttranscriptional gene expression regulation caused by double stranded RNA molecules. This is one of the most willingly used experimental techniques. It is used in experiments designed to explore gene function, in creation cellular models of diseases or in genetic therapy design. Among RNAi technology reagents are siRNA (short interfering RNA) and vector reagents shRNA (short hairpin RNA) and sh-miR (sh- microRNA). RNAi reagents are integrated into endogenous RNAi pathway on different phases. sh-miR reagents are designed in the way to be similar to endogenous miRNA precursors (pri-miRNA), because it is important feature to be recognized by proteins involved in microRNA (miRNA) biogenesis. shRNA or sh-miR reagents are introduced into cells with usage of genetic vectors – plasmids or viral vectors to ensure intracellular expression. They have huge therapeutic potential beacuse of stable reagent production in cells what enables one time therapeutic dosage. Usage of synthetic siRNA reagents requires often dosage, what makes therapy more difficult, especially in case of Central Nervous System Diseases.
shRNA or sh-miR reagents are introduced into cells with usage of genetic vectors – plasmids or viral vectors to ensure intracellular expression. They have huge therapeutic potential beacuse of stable reagent production in cells what enables one time therapeutic dosage. Usage of synthetic siRNA reagents requires often dosage, what makes therapy more difficult, especially in case of Central Nervous System Diseases.
 It was recently shown that RNAi reagents introduces into cells can trigger unwanted non-specific effects, e.g. can interact with different transcripts besides the desired one (so called off-target effect) or induce immunological effect. Because in processing of sh-miR reagents endogenous endonucleasis Drosha and Dicer are involved, in case of high expression of reagent they can saturate those proteins as well as Exportin-5 transport protein responsible for export of sh-miR from nucleus. The saturation can influence endogenous RNAi mechanism. It was shown that sh-miR reagents influence less endogenous RNAi pathway, because of lower level of produced double-stranded RNA (dsRNA), preserving efficiency of translation silencing. They also show longer effect than different types of RNAi reagents. Because of described features sh-miR reagents seems to be good choice when designing efficient and non-toxic RNAi reagents.
sh-miR Designer software was designed sh-miR Designer is a software aimed for fast and efficient design of effective RNA interference (RNAi) reagents - sh-miRs, also known as artificial miRNAs. sh-miRs are RNA particles whose structure is based on miRNA precursor pri-miRNA, but sequence interacting with transcript is changed depending on research purpose. Until now scientists have to use multiple unconnected software to design single sh-miR molecule. It is time consuming process, what limits number of designed molecules for biological testing. There is no such software, which includes all important features as molecule spatial structure. Maintenance of structure of pri-miRNA is very important to enable cellular processing and therefore ensure functionality of artificial particles. This interdisciplinary project includes computer sciences with biomedical science. sh-miRs delivered to cells on genetic vectors - plasmids or viral vectors - enter natural RNAi pathway and silence target mRNA. They can be used in genetic therapies and basic biomedical research.
It was recently shown that RNAi reagents introduces into cells can trigger unwanted non-specific effects, e.g. can interact with different transcripts besides the desired one (so called off-target effect) or induce immunological effect. Because in processing of sh-miR reagents endogenous endonucleasis Drosha and Dicer are involved, in case of high expression of reagent they can saturate those proteins as well as Exportin-5 transport protein responsible for export of sh-miR from nucleus. The saturation can influence endogenous RNAi mechanism. It was shown that sh-miR reagents influence less endogenous RNAi pathway, because of lower level of produced double-stranded RNA (dsRNA), preserving efficiency of translation silencing. They also show longer effect than different types of RNAi reagents. Because of described features sh-miR reagents seems to be good choice when designing efficient and non-toxic RNAi reagents.
sh-miR Designer software was designed sh-miR Designer is a software aimed for fast and efficient design of effective RNA interference (RNAi) reagents - sh-miRs, also known as artificial miRNAs. sh-miRs are RNA particles whose structure is based on miRNA precursor pri-miRNA, but sequence interacting with transcript is changed depending on research purpose. Until now scientists have to use multiple unconnected software to design single sh-miR molecule. It is time consuming process, what limits number of designed molecules for biological testing. There is no such software, which includes all important features as molecule spatial structure. Maintenance of structure of pri-miRNA is very important to enable cellular processing and therefore ensure functionality of artificial particles. This interdisciplinary project includes computer sciences with biomedical science. sh-miRs delivered to cells on genetic vectors - plasmids or viral vectors - enter natural RNAi pathway and silence target mRNA. They can be used in genetic therapies and basic biomedical research.
Standard PartsWe are creating a particle which can be inserted into plasmid built by Standard parts from Registry of Standard Biological Parts. The sh-miR molecules are inserted into plasmid backbone downstream chosen promoter. You can add any regulatory sequence you want or maybe a fragment coding fluorescent protein to see if the transfection was succesfull. The molecules are fully compatibile with Standard Biological Parts - what is important for users from iGEM especially :).
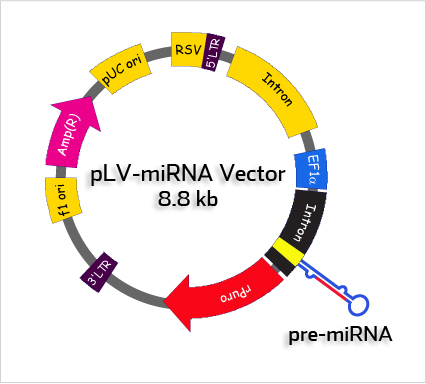 picture from http://biosettia.com/
picture from http://biosettia.com/
User Guide
There are two suplementary versions of our software. First one (based on last year's software sh-miR v1.0) enable to design sh-miR molecules based on siRNA sequence and the second design sh-miR molecules when NCBI transcript number (NM_...) is provided. In the second version the user also can set GC content, maximum off-target and immunostimulatory properties. The user can also set which pri-miRNA should be used to design sh-miR (or all available in our miRNA database).
Please follow video where you can see how to use web version of the software
and short description how to use client version of the software
Both versions are well documented on github and readthedocs (see below)
Usage via Web server
go to our website shmir.wmi.amu.edu.pl
Technology
The application was built with Python 2.7. It is divided into 3 parts:
RESTful API - the “heart” of our software - it performs all calculations. Our application is built on CentOS (Linux) operating system. To maintain better compatibility with our server we use Vagrant to create Virtual Machine and Puppet to manage infrastructure of our project. All you need to do is: type into console “vagrant up” and after a while everything is ready to work. This subproject is written with following technologies/frameworks/libraries: Flask, Flask-Cache, SqlAlchemy, PostgreSQL, Celery, Flower, Rabbitmq, Redis, BioPython, mfold (lightweight), uwsgi, sphinx, Python testing tools (mock, nose, unittest).
Console client - simple console application which communicates with API using requests library. It has 3 options: mfold (folding structure), from_sirna (create sh-miR from siRNA) and from_transcript (create sh-miR from transcript name).
Website - application written in Django framework. It enables you to use the API via graphical interface. You can also create your own account and save the history of your requests for later usage.
API documentationHere you can find documentation from code: API: shmir-api.rtfd.org Client: shmir-client.rtfd.org Webpage: shmir-designer.rtfd.orgAPI for developersHere you can find api for developers: https://github.com/Nozdi/shmirReleases (re-use of 2013iGEM software)sh-miR designer v1.0First version of the program presented on iGEM 2013 by our team. The user have to provide siRNA sequence (one or both strands). The miRNA database includes 5 miRNAs.sh-miR designer v1.1Strand discrimination functionality was added based on miRbase. extended miRNA databasesh-miR designer v2.0siRNA prediction algorithm was added to functionality (the input is NCBI transcript number) user can choose GC content of siRNA molecule, maximal off-target, if he/she wants immunostimulatory sequences inside the molecule and to choose if he/she wants to include all or chosen scaffold extended miRNA database What we are working on: strand discrimination based on therodynamics of siRNA ends; preparing construct with flanking restriction enzymes providing plasmids with parts from standard partsUnit Tests
In our application we are using unit tests to test alghoritms, functions and database.
We are testing inputs and outputs:
def test_validate_gc_content(self):
is_in_range = validate_gc_content('ACGT', 40, 60)
self.assertTrue(is_in_range)
Caught errors:
def multiple_input(self, data, error):
for strand in data:
with self.assertRaises(errors.InputException) as err:
validators.check_input(strand)
self.assertEqual(error, str(err.exception))
Database models:
def test_get_and_put_to_db(self):
new_obj = self.TestModel(id=153)
self.put_to_db(new_obj)
objs = self.get_all(self.TestModel)
self.assertEqual(len(objs), 1)
We are using Mock library to test our functions which are using external API’s to create fake response from server and return them into our function:
@patch.object(ncbi_api.Entrez, 'esearch')
@patch.object(ncbi_api.Entrez, 'efetch')
def test_get_data(self, mock_efetch, mock_esearch):
ids = [69]
mock_read = Mock()
mock_read.read.return_value = json.dumps({'esearchresult': {'idlist': ids}})
mock_esearch.return_value = mock_read
ncbi_api.get_data(self.GOOD_MRNA)
mock_esearch.assert_called_with(
term='NM_001128164.1', db=self.DATA_BASE, retmode='json'
)
mock_efetch.assert_called_once_with(
db=self.DATA_BASE, id=ids, rettype='fasta', retmode='text'
)
Validationold version shmiR v1.0 (average time)real 0m6.854s user 0m0.063s sys 0m0.027snew version shmiR v1.1 (average times)real 0m3.271s user 0m0.155s sys 0m0.041sDesigned sh-miRfor $ ./shmir_client.py from_transcript NM_000546.5 1: backbone: miR-31, score: 106, sequence: ACATGACGGAGGTTGTGAG pdf: ./results/transcript/e9e8b9a2-7e3b-4b24-b26a-633332a3f6b2/miR-31/7270b2d2-0aca-472b-853e-5a9631164d46 result: CATAACAACGAAGAGGGATGGTATTGCTCCTGTAACTCGGAACTGGAGAGGACATGACGGAGGTTGTGAGCGTTGAACTGGGAACCCTCACAACCTCCGTCA TGTCTCTCCAGTTCCGAGTTACAGGAGCAATACCATCCCTCTTCGTTGTTATGCTTTCCTGTCTGACAGCAGCTTGGCTACCTCCGTCCTGTTCCTCCTTGTCTT 2: backbone: miR-31, score: 106, sequence: ATGTTTTGTAGAGATGGGG pdf: ./results/transcript/e9e8b9a2-7e3b-4b24-b26a-633332a3f6b2/miR-31/56232916-2649-4eb4-9970-c0b0a08ce64e result: CATAACAACGAAGAGGGATGGTATTGCTCCTGTAACTCGGAACTGGAGAGGATGTTTTGTAGAGATGGGGCGTTGAACTGGGAACCCCCCATCTCTACA AAACATCTCTCCAGTTCCGAGTTACAGGAGCAATACCATCCCTCTTCGTTGTTATGCTTTCCTGTCTGACAGCAGCTTGGCTACCTCCGTCCTGTTCCTCCTTGTCTT 3: backbone: miR-31, score: 106, sequence: ATTACATGGGGTCTAGAAC pdf: ./results/transcript/e9e8b9a2-7e3b-4b24-b26a-633332a3f6b2/miR-31/37cd84aa-362c-484d-8681-929043cc9c61 result: CATAACAACGAAGAGGGATGGTATTGCTCCTGTAACTCGGAACTGGAGAGGATTACATGGGGTCTAGAACCGTTGAACTGGGAACCGTTCTAG ACCCCATGTAATCTCTCCAGTTCCGAGTTACAGGAGCAATACCATCCCTCTTCGTTGTTATGCTTTCCTGTCTGACAGCAGCTTGGCTACCTCCGTCCTGTTCCTCCTTGTCTT 4: backbone: miR-31, score: 106, sequence: ACCATCATCACACTGGAAG pdf: ./results/transcript/e9e8b9a2-7e3b-4b24-b26a-633332a3f6b2/miR-31/50b763f6-2193-4743-96c0-61601880e574 result: CATAACAACGAAGAGGGATGGTATTGCTCCTGTAACTCGGAACTGGAGAGGACCATCATCACACTGGAAGCGTTGAACTGGGAACCCTT CCAGTGTGATGATGGTCTCTCCAGTTCCGAGTTACAGGAGCAATACCATCCCTCTTCGTTGTTATGCTTTCCTGTCTGACAGCAGCTTGGCTACCTCCGTCCTGTTCCTCCTTGTCTT 5: backbone: miR-31, score: 106, sequence: ATCTCACCCCATCCCACAC pdf: ./results/transcript/e9e8b9a2-7e3b-4b24-b26a-633332a3f6b2/miR-31/019ce915-0282-4d83-9538-93c36c790fa8 result: CATAACAACGAAGAGGGATGGTATTGCTCCTGTAACTCGGAACTGGAGAGGATCTCACCCCATCCCACACCGTTGAAC TGGGAACCGTGTGGGATGGGGTGAGATCTCTCCAGTTCCGAGTTACAGGAGCAATACCATCCCTCTTCGTTGTTATGCTTTCCTGTCTGACAGCAGCTTGGCTACCTCCGTCCTGTTCCTCCTTGTCTT see example pdf with structure link During sh-miR designer development, colleagues from befriended lab used our software, so we had constant feedback from potential users, who by the way, have nothing to do with bioinformatics. So it's really hard to satisfy their strong need for easy interface. We also had significant feedback from our survey (see Modeling tab), so we knew what we should emphasise on during develoment.Test cases1) ATXN3 - therapeutic sh-miR against SCA3 disease transcript number: NM_001164782.1 GC content: default Off-target: 10 scaffold: ALL immunostimulatory: NO 2) P54 - therapeutic sh-miR against cancer transcript number: NM_000546.5 GC content: default Off-target: 10 scaffold: ALL immunostimulatory: YES 3) biological function analysis, PARP1, we have miR-30a in our lab transcript number: NM_001618.3 GC content: default Off-target: 0 scaffold: miR-30a immunostimulatory: no_difference (default) 4) biological function, PARP1, we have miR-30a in our laboratory and have immune cells transcript number: NM_001618.3 GC content: default Off-target: 0 scaffold: miR-30a immunostimulatory: NO 5) biological function, PARP1 and have immune cells transcript number: NM_001618.3 GC content: default Off-target: 0 scaffold: ALL immunostimulatory: NOReferences
1. Boudreau RL, Monteys AM, Davidson BL. Minimizing variables among hairpin-based RNAi
vectors reveals the potency of shRNAs. RNA. 2008 Sep;14(9):1834-44. Epub 2008 Aug 12.
2. Chang K, Elledge SJ, Hannon GJ. Lessons from Nature: microRNA-based shRNA libraries. Nat
Methods. 2006 Sep;3(9):707-14.
3. Dow LE, Premsrirut PK, Zuber J, Fellmann C, McJunkin K, Miething C, Park Y, Dickins RA,
Hannon GJ, Lowe SW. A pipeline for the generation of shRNA transgenic mice. Nat Protoc.
2012 Feb 2;7(2):374-93. doi: 10.1038/nprot.2011.446.
4. Fahim M, Larkin PJ. Designing Effective amiRNA and Multimeric amiRNA Against Plant
Viruses. Methods Mol Biol. 2013;942:357-77. doi: 10.1007/978-1-62703-119-6_19.
5. Fan ZD, Zhang L, Shi Z, Gan XB, Gao XY, Zhu GQ. Artificial microRNA interference targeting
AT(1a) receptors in paraventricular nucleus attenuates hypertension in rats. Gene Ther. 2012
Aug;19(8):810-7. doi: 10.1038/gt.2011.145. Epub 2011 Sep 29.
6. Fellmann C, Zuber J, McJunkin K, Chang K, Malone CD, Dickins RA, Xu Q, Hengartner MO,
Elledge SJ, Hannon GJ, Lowe SW. Functional identification of optimized RNAi triggers using a
massively parallel sensor assay. Mol Cell. 2011 Mar 18;41(6):733-46. Epub 2011 Feb 25.
7. Maczuga P, Lubelski J, van Logtenstein R, Borel F, Blits B, Fakkert E, Costessi A, Butler D, van
Deventer S, Petry H, Koornneef A, Konstantinova P. Embedding siRNA sequences targeting
Apolipoprotein B100 in shRNA and miRNA scaffolds results in differential processing and in
vivo efficacy. Mol Ther. 2012 Oct 23. doi: 10.1038/mt.2012.160. [Epub ahead of print]
8. Maczuga P, Koornneef A, Borel F, Petry H, van Deventer S, Ritsema T, Konstantinova P.
Optimization and comparison of knockdown efficacy between polymerase II expressed
shRNA and artificial miRNA targeting luciferase and Apolipoprotein B100. BMC
Biotechnol. 2012 Jul 24;12:42. doi: 10.1186/1472-6750-12-42.
9. Matveeva OV, Nazipova NN, Ogurtsov AY, Shabalina SA. Optimized models for design of
efficient miR30-based shRNAs. Front Genet. 2012;3:163. Epub 2012 Aug 29.
10. McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, Gilmore BL, Burstein
H, Peluso RW, Polisky B, Carter BJ, Davidson BL. Artificial miRNAs mitigate shRNA-mediated
toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci
U S A. 2008 Apr 15;105(15):5868-73. Epub 2008 Apr 8.
11. Schwab R, Ossowski S, Warthmann N, Weigel D. Directed gene silencing with artificial
microRNAs. Methods Mol Biol. 2010;592:71-88.
12. Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach
MR, Sheth N, Bradshaw J, Burchard J, Kulkarni A, Cavet G,Sachidanandam R, McCombie WR, Cleary MA, Elledge SJ, Hannon GJ. Second-generation shRNA libraries
covering the mouse and human genomes. Nat Genet. 2005 Nov;37(11):1281-8. Epub 2005
Oct 2.
13. Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for
single-copy polymerase II-regulated RNA interference in mammalian cells. Proc Natl Acad Sci
U S A. 2005 Sep 13;102(37):13212-7. Epub 2005 Sep 1.
14. Wang X, Yang Y, Zhou J, Yu C, Cheng Y, Yan C, Chen J.Two-step method for constructing
Arabidopsis artificial microRNA vectors. Biotechnol Lett. 2012 Jul;34(7):1343-9. Epub 2012
Mar 22.
15. Warthmann N, Ossowski S, Schwab R, Weigel D. Artificial MicroRNAs for Specific Gene
Silencing in Rice. Methods Mol Biol. 2013;956:131-49. doi: 10.1007/978-1-62703-194-3_11.
16. Zeng Y, Cullen BR. Efficient processing of primary microRNA hairpins by Drosha requires
flanking nonstructured RNA sequences. J Biol Chem. 2005 Jul 29;280(30):27595-603. Epub
2005 Jun 1.
17. Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the
expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002 Jun;9(6):1327-
33
Judging Form (in bold what we did to achieve gold medal)Bronze. The following 4 goals must be achieved:
|
|||||||||||||||
 "
"


 users' history:
users' history:


