Team:SUSTC-Shenzhen/Project/Plasmid
From 2014.igem.org
(Created page with "{{SUSTC-Shenzhen/removeStyles}} {{SUSTC-Shenzhen/themeCss}} {{:Team:SUSTC-Shenzhen/templates/nav-template}} {{:Team:SUSTC-Shenzhen/templates/page-header| title=Plasmid| ...") |
(→gRNA-mCherry-NLS-UAS plasmids) |
||
| (8 intermediate revisions not shown) | |||
| Line 10: | Line 10: | ||
Here are the full list of plasmids we've used and constructed and their names and maps have universality. | Here are the full list of plasmids we've used and constructed and their names and maps have universality. | ||
| + | |||
| + | =gRNA-mCherry-NLS-UAS plasmids= | ||
| + | {{SUSTC-Image|wiki/images/8/87/SUSTC-Shenzhen-gRNAforEGFP.png}} | ||
| + | |||
| + | =Plasmids from Germany= | ||
| + | pSW55-GD5 (partial) | ||
| + | Uherek et al. 1998 | ||
| + | Brief introduction: | ||
| + | pSW55-GD5 encodes the chimeric fusion protein GD5 working as non-viral vector that can transfer specific target gene into specific target cells. | ||
| + | |||
| + | Contains: | ||
| + | partial sequence (coding sequence) | ||
| + | beginning with NdeI site (CATATG) | ||
| + | vector backbone (not included): | ||
| + | pFLAG-1, ompA signal peptide removed | ||
| + | nt 4-1854: coding sequence GD5 fusion protein | ||
| + | nt 7-30 FLAG peptide | ||
| + | nt 37-54: 6 x His tag | ||
| + | nt 55-492: GAL4 residues 2 to 147 (DNA binding domain) | ||
| + | nt 505-1071 DT-B translocation domain | ||
| + | nt 1093-1815: scFv(FRP5) (anti-ErbB2) | ||
| + | [[File:SUSTC-Shenzhen-pSW55-GD5(coding_sequence).jpg|thumb|Fig.1 GD5]] | ||
| + | |||
| + | Click the linking at the following for detailed information | ||
| + | https://2014.igem.org/Team:SUSTC-Shenzhen/Notebook/A-B_Toxin/Purify | ||
| + | lick the linking at the following for summarized information | ||
| + | https://2014.igem.org/Team:SUSTC-Shenzhen/Project/A-B_toxin#Construction | ||
| + | |||
| + | |||
| + | |||
| + | pWF47-TEG (partial) | ||
| + | Fominaya et al. 1998 | ||
| + | (corrected 2014-07-31) | ||
| + | Brief introduction: | ||
| + | pWF47-TEG encodes the chimeric fusion protein TEG working as non-viral vector that can transfer specific target gene into specific target cells. | ||
| + | |||
| + | partial sequence (coding sequence) | ||
| + | beginning with second half of NdeI site (catATG) | ||
| + | vector backbone (not included): pFLAG-1 | ||
| + | nt 1-1089: coding sequence TEG fusion protein | ||
| + | nt 1-63: ompA signal peptide (pFLAG-1) | ||
| + | nt 64-87: FLAG peptide (pFLAG-1) | ||
| + | nt 100-249: hTGFalpha residues 40 to 89 (mature form) | ||
| + | nt 259-276: 6 x His tag | ||
| + | nt 280-615: ETA domain II | ||
| + | nt 628-1065: GAL4 residues 2 to 147 (DNA binding domain) | ||
| + | nt 1078-1089: KDEL ER retention sequence | ||
| + | |||
| + | [[File:SUSTC-Shenzhen-PWF47-TEG(coding_sequence).jpg|thumb|Fig.2 TEG]] | ||
| + | |||
| + | Click the linking at the following for detailed information | ||
| + | https://2014.igem.org/Team:SUSTC-Shenzhen/Notebook/A-B_Toxin/Purify | ||
| + | lick the linking at the following for summarized information | ||
| + | https://2014.igem.org/Team:SUSTC-Shenzhen/Project/A-B_toxin#Construction | ||
| + | |||
| + | =Target-sequence= | ||
| + | |||
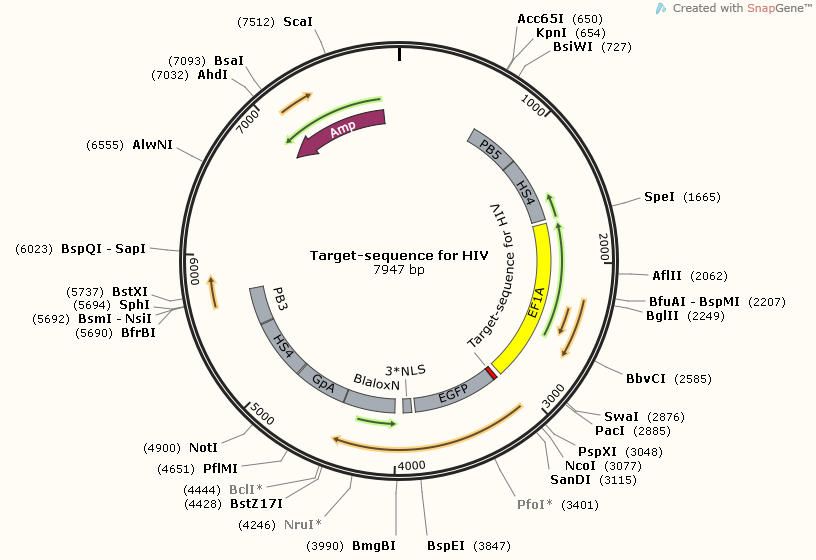
| + | [https://2014.igem.org/Team:SUSTC-Shenzhen/Project/gRNA-binding-target-sequence_Plasmids_Design_and_Construction '''Target-sequence_for_HIV'''] | ||
| + | [[File:Target-sequence_for_HIV_Map.png]] | ||
| + | |||
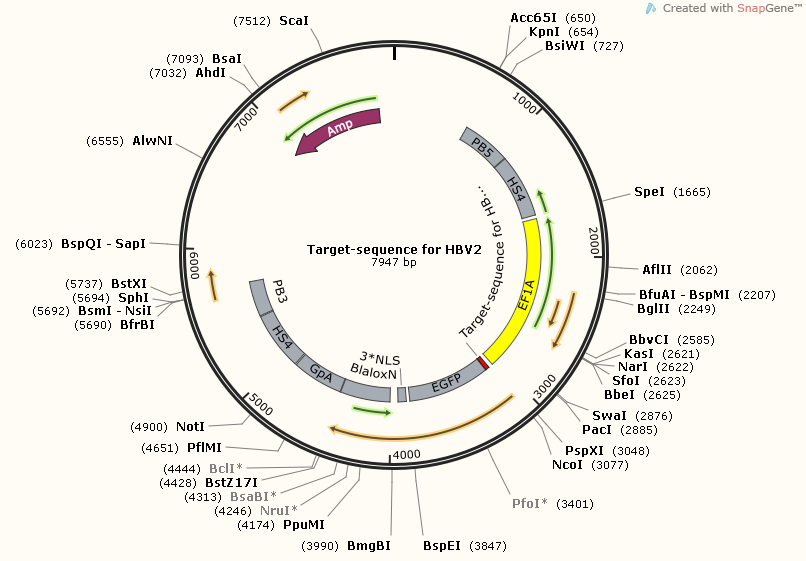
| + | [https://2014.igem.org/Team:SUSTC-Shenzhen/Project/gRNA-binding-target-sequence_Plasmids_Design_and_Construction '''Target-sequence_for_HBV2'''] | ||
| + | [[File:Target-sequence_for_HBV2_Map.png]] | ||
| + | |||
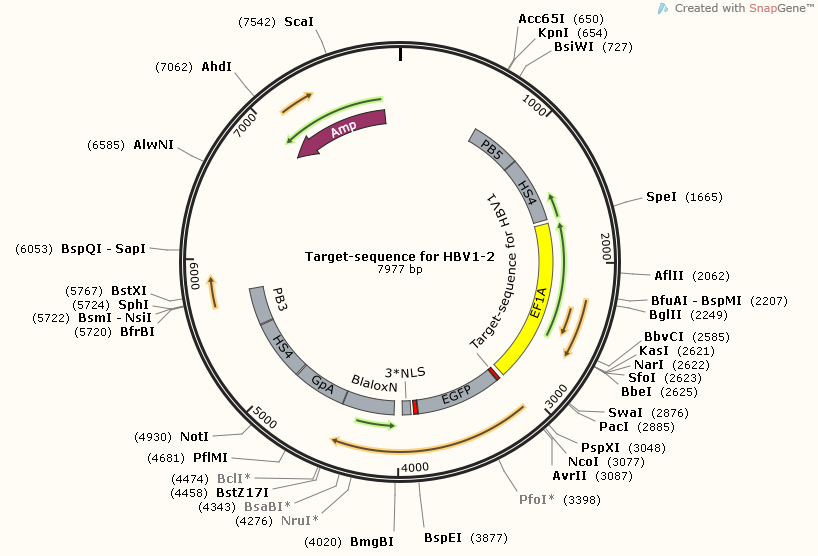
| + | [https://2014.igem.org/Team:SUSTC-Shenzhen/Project/gRNA-binding-target-sequence_Plasmids_Design_and_Construction '''Target-sequence_for_HBV1-2'''] | ||
| + | [[File:Target-sequence_for_HBV1-2_Map.png]] | ||
{{SUSTC-Shenzhen/main-content-end}} | {{SUSTC-Shenzhen/main-content-end}} | ||
{{SUSTC-Shenzhen/wiki-footer}} | {{SUSTC-Shenzhen/wiki-footer}} | ||
{{SUSTC-Shenzhen/themeJs}} | {{SUSTC-Shenzhen/themeJs}} | ||
Latest revision as of 03:55, 18 October 2014
Plasmid
Full list of plasmid we used and constructed
Here are the full list of plasmids we've used and constructed and their names and maps have universality.
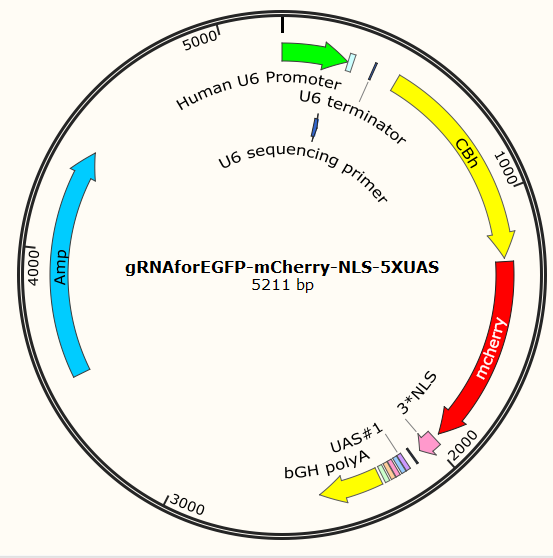
gRNA-mCherry-NLS-UAS plasmids
Plasmids from Germany
pSW55-GD5 (partial) Uherek et al. 1998 Brief introduction: pSW55-GD5 encodes the chimeric fusion protein GD5 working as non-viral vector that can transfer specific target gene into specific target cells.
Contains: partial sequence (coding sequence) beginning with NdeI site (CATATG) vector backbone (not included): pFLAG-1, ompA signal peptide removed nt 4-1854: coding sequence GD5 fusion protein nt 7-30 FLAG peptide nt 37-54: 6 x His tag nt 55-492: GAL4 residues 2 to 147 (DNA binding domain) nt 505-1071 DT-B translocation domain nt 1093-1815: scFv(FRP5) (anti-ErbB2)
Click the linking at the following for detailed information https://2014.igem.org/Team:SUSTC-Shenzhen/Notebook/A-B_Toxin/Purify lick the linking at the following for summarized information https://2014.igem.org/Team:SUSTC-Shenzhen/Project/A-B_toxin#Construction
pWF47-TEG (partial) Fominaya et al. 1998 (corrected 2014-07-31) Brief introduction: pWF47-TEG encodes the chimeric fusion protein TEG working as non-viral vector that can transfer specific target gene into specific target cells.
partial sequence (coding sequence) beginning with second half of NdeI site (catATG) vector backbone (not included): pFLAG-1 nt 1-1089: coding sequence TEG fusion protein nt 1-63: ompA signal peptide (pFLAG-1) nt 64-87: FLAG peptide (pFLAG-1) nt 100-249: hTGFalpha residues 40 to 89 (mature form) nt 259-276: 6 x His tag nt 280-615: ETA domain II nt 628-1065: GAL4 residues 2 to 147 (DNA binding domain) nt 1078-1089: KDEL ER retention sequence
Click the linking at the following for detailed information https://2014.igem.org/Team:SUSTC-Shenzhen/Notebook/A-B_Toxin/Purify lick the linking at the following for summarized information https://2014.igem.org/Team:SUSTC-Shenzhen/Project/A-B_toxin#Construction
Target-sequence
 "
"