Team:Oxford/notebook
From 2014.igem.org
| Line 28: | Line 28: | ||
<BR> | <BR> | ||
| + | </html> | ||
[[File:Oxfordigem_B1-1.png|centre]]<BR> | [[File:Oxfordigem_B1-1.png|centre]]<BR> | ||
| + | <html> | ||
We ran these 4 PCR reactions as follows using the NEB Q5 Protocol: | We ran these 4 PCR reactions as follows using the NEB Q5 Protocol: | ||
<BR> | <BR> | ||
| + | </html> | ||
[[File:Oxfordigem_B1-2.png |centre]]<BR> | [[File:Oxfordigem_B1-2.png |centre]]<BR> | ||
| - | + | <html> | |
<h1>Part C</h1> | <h1>Part C</h1> | ||
Revision as of 14:49, 21 July 2014
_NOTOC__
Lab Book
Part A
Part B
Week 1
Tasks completed:
Swapping resistance gene for plasmid pME6010 from tetracycline to kanamycin.
Methods:
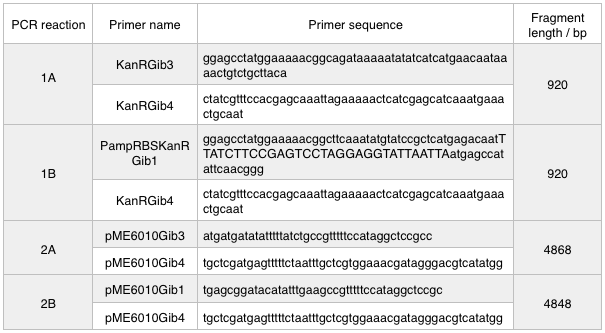
First, we designed forward and reverse primers for KanR (with native RSB and promoter) which we isolated from plasmid pCM66. The 5’ ends were complementary to the insert region of pME6010 (reaction 1).
We then designed forward and reverse primers to amplify the pME6010 backbone (reaction 2).
Further to this we redesigned each set of primers to incorporate an ampicillin promoter and optimised RBS (https://salis.psu.edu/software/) (B reactions) instead of the native promoter region (A reactions).
The designed plasmids were as follows:
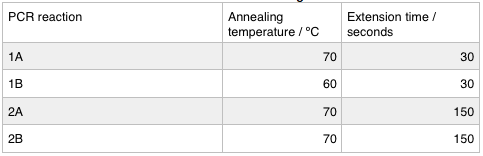
We ran these 4 PCR reactions as follows using the NEB Q5 Protocol:
 "
"