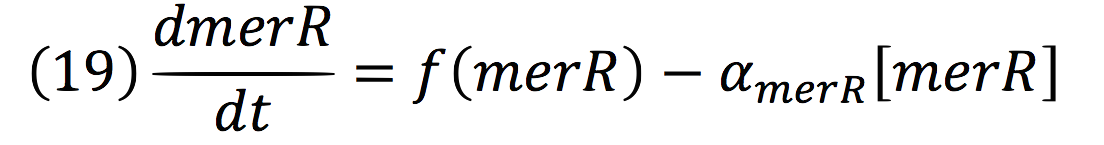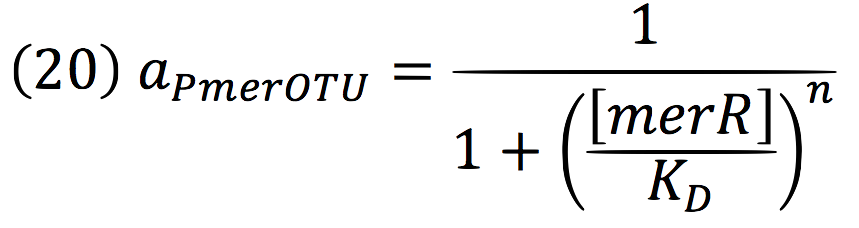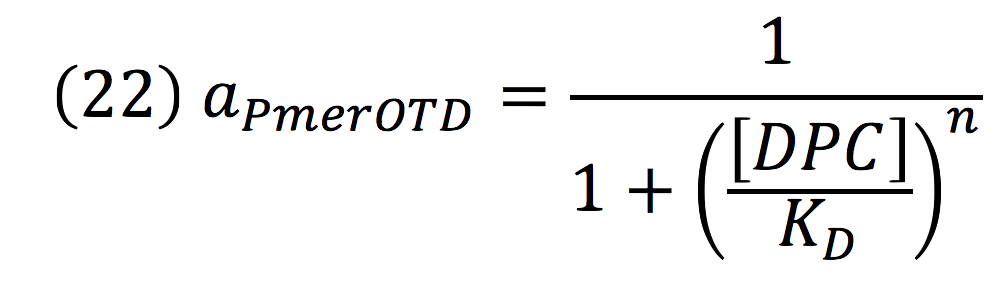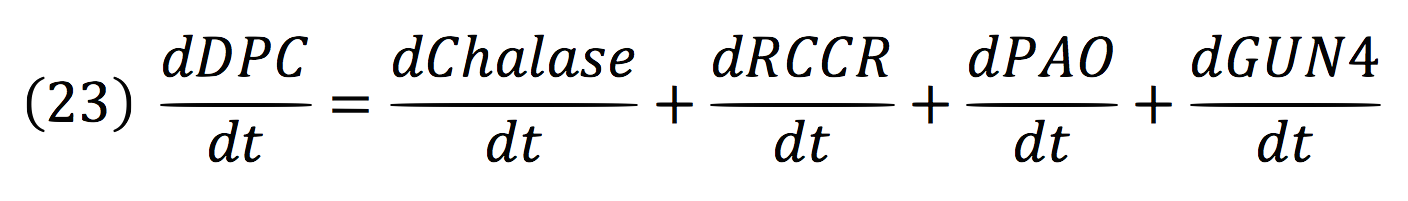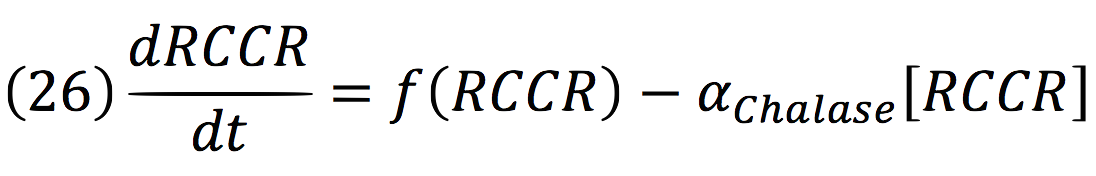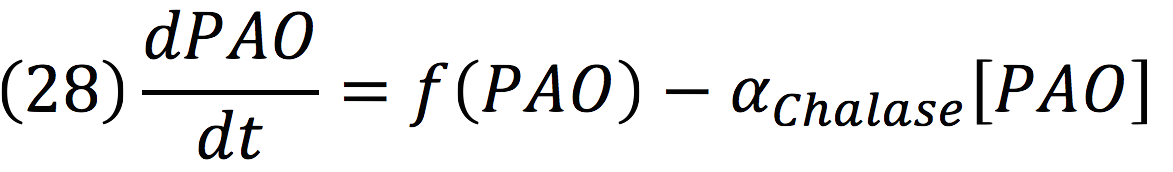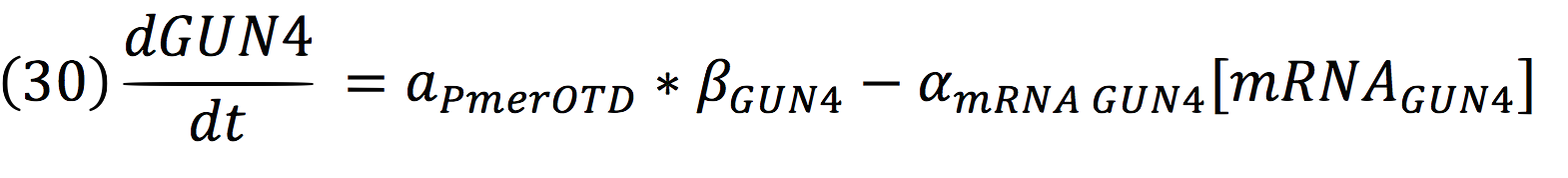Team:BIOSINT Mexico/Sensor
From 2014.igem.org
(→Inducible promoter by Hg(II)) |
(→Biosynthesis and Breakdown) |
||
| Line 21: | Line 21: | ||
===Biosynthesis and Breakdown=== | ===Biosynthesis and Breakdown=== | ||
| - | |||
The first phase of the chlorophyll biosynthesis starts with the glutamic acid, after nine chemical steps this amino acid produces a four ring structure, called protoporphyrin IX. A molecule of magnesium is added to the ring structure by the magnesium chelatase, through two more steps this is converted in monovinyl protochlorophyllide and is reduce to chlorophyllide a. by the enzyme protochlorophyllide oxidoreductase (POR). The chlorophyllide a. is transformed in darker green chlorophyll by the chlorophyll synthetase enzyme, this add a 20 carbon phytol tail. | The first phase of the chlorophyll biosynthesis starts with the glutamic acid, after nine chemical steps this amino acid produces a four ring structure, called protoporphyrin IX. A molecule of magnesium is added to the ring structure by the magnesium chelatase, through two more steps this is converted in monovinyl protochlorophyllide and is reduce to chlorophyllide a. by the enzyme protochlorophyllide oxidoreductase (POR). The chlorophyllide a. is transformed in darker green chlorophyll by the chlorophyll synthetase enzyme, this add a 20 carbon phytol tail. | ||
The chlorophyll pathway degradation is an important catabolic process for the senescence of the leaf. This breakdown pathway starts with the chlorophyllase enzyme, which removes the hydrophobic twenty carbon phytol tail from the chlorophyll. As the synthetic pathway, the chlorophyll turn into the light green molecule called chlorophyllide. This molecule is converted to pheophorbide a. by the magnesium dechelatase enzyme at removing the magnesium and the red chlorophyll catabolite (RCC) is formed aside pheophorbide a oxygenase. Then the RCC reductase produces fluorescent chlorophyll catabolite (FCC). The FCC goes through different steps and its converted into nonfluorescent chlorophyll catabolites. | The chlorophyll pathway degradation is an important catabolic process for the senescence of the leaf. This breakdown pathway starts with the chlorophyllase enzyme, which removes the hydrophobic twenty carbon phytol tail from the chlorophyll. As the synthetic pathway, the chlorophyll turn into the light green molecule called chlorophyllide. This molecule is converted to pheophorbide a. by the magnesium dechelatase enzyme at removing the magnesium and the red chlorophyll catabolite (RCC) is formed aside pheophorbide a oxygenase. Then the RCC reductase produces fluorescent chlorophyll catabolite (FCC). The FCC goes through different steps and its converted into nonfluorescent chlorophyll catabolites. | ||
| - | |||
===De-greening=== | ===De-greening=== | ||
Revision as of 02:48, 18 October 2014
Modular Sensor
Description
There has been an increasing necessity to implement new, efficient and inexpensive techniques for the identification of biological and chemical agents that contaminate the environment, one of the most developed strategies for solving this trouble is the use of biosensors. A reporter device must be an easily detectable mechanism for sensing a specific substance of interest, this reporter needs to have a monitoring and a resettable capacity.
In this project the target pollutant that will induce gene expression of the biological sensor is the mercury, this is based on the loss of green pigmentation in Arabidopsis thaliana. A substantially faster loss of chlorophyll is needed if is used as a reporter system for a plant sentinel. Chlorophyll loss in plants is normally a slow process that occurs during the complex mechanism of senescence. The half-life of chlorophyll has been estimated to be 2–5 days for relatively mature and fully greened leaves (Stobart and Hendry, 1984), visual perception of chlorophyll loss in leaves can take longer.
The process for whitening the plant requires following the method explained below and takes around 48 hours in order to see a complete change in color. In order to accomplish this, it is necessary to use specific enzymes which degrades the chlorophyll that is already in the plant and also a doubled stranded RNA which inhibits the production of an important molecule involved in the production of chlorophyll (Medford, J. et al , 2006).
Biosynthesis and Breakdown
The first phase of the chlorophyll biosynthesis starts with the glutamic acid, after nine chemical steps this amino acid produces a four ring structure, called protoporphyrin IX. A molecule of magnesium is added to the ring structure by the magnesium chelatase, through two more steps this is converted in monovinyl protochlorophyllide and is reduce to chlorophyllide a. by the enzyme protochlorophyllide oxidoreductase (POR). The chlorophyllide a. is transformed in darker green chlorophyll by the chlorophyll synthetase enzyme, this add a 20 carbon phytol tail.
The chlorophyll pathway degradation is an important catabolic process for the senescence of the leaf. This breakdown pathway starts with the chlorophyllase enzyme, which removes the hydrophobic twenty carbon phytol tail from the chlorophyll. As the synthetic pathway, the chlorophyll turn into the light green molecule called chlorophyllide. This molecule is converted to pheophorbide a. by the magnesium dechelatase enzyme at removing the magnesium and the red chlorophyll catabolite (RCC) is formed aside pheophorbide a oxygenase. Then the RCC reductase produces fluorescent chlorophyll catabolite (FCC). The FCC goes through different steps and its converted into nonfluorescent chlorophyll catabolites.
De-greening
Since the chlorophyll degradation is a slow process, a synthetic system that removes and degrades chlorophyll in a fast and efficient way is needed. For this reason we used a de-greening circuit, that allows the regulation of the chlorophyll breakdown and the response to a specific input. Our system detects heavy metals, and in this case we used methylmercury, which is transcriptionally linked to the de-greening system.
One of the solutions for accelerating the breakdown is an inducible gene silencing system using interference RNA. The silencing gene used was phytoene desaturase( PDS). This enzymes produced by PDS blocks carotenoids synthesis, culminating in a photobleaching phenotype given by the photo-oxidation of chlorophylls.
If a characteristic white phenotype is going to be used to identify methylmercury presence, it must be much faster than previous RNAi circuits. Medford, J et al (2006) proposed the combination of the following regulatory circuits for de-greening:
a) Stop synthesis circuit: induction of diRNA to reduce protochlorophyllide oxidoreductase (POR) or diRNA to lessen GENOMES UNCOUPLED 4 (GUN4). For the project we only use GUN4, this is a single copy gene that regulates chlorophyll biosynthesis by activating magnesium chelatase, a key enzyme complex that produces magnesium protoporphyrin IX, basic structure for chlorophyll. With a double-stranded interfering RNA construct designed for each one of them and placed under the control of certain promoter, A. thaliana demonstrates chlorophyll loss. (Antunes et. al 2006).
b) Initiate breakdown circuit: induction of chlorophyllase (CHLASE) and red chlorophyll catabolite reductase (RCCR) or CHLASE and pheophorbide a oxygenase (PAO).Chlorophyll breakdown involves a series of enzymatic steps. Key processes are the hydrophobic tail removal by CHLASE, red chlorophyll catabolite reductase (RCCR) and porphyrin ring cleavage by PAO.
By combining both processes the de-greening is easily recognized within 24-48 hrs of induction. Also we added two more miRNA for the de-greening circuit: one of the miRNA it's for the RBS and the other it's for the IRES, both of them are in the system of the chlorophyll breakdown.
Inducible promoter by Hg(II)
For the activation of the degreening device the use of an inducible promoter is needed. Since the project is about bioremediation of methylmercury, we applied the double promoter of the mercury resistance operon of the transposon Tn21. These are two overlapping divergent oriented promoters, the first one regulates the MerR protein and it’s called PR, the second it’s called PTPCAD and it regulates the gene transcription. The set of these promoters it’s called MerOP .
The substance that activates and regulates merOP promoter is the Hg(II). The PTPCAD expression is repressed in the absence of Hg (II) by the merR gene, this merR product is used as a regulator. Meanwhile the expression of PR doesn’t change in the presence or absence of this molecule. MerR always represses its own expression so it doesn’t matter of there is presence of Hg(II).
MerR functions as a homodimer and it’s always bound to the MerOP. This has an extremely affinity for Hg(II) due to its unique ability to coordinate that molecule in a trigonal planar configuration. MerR has three cysteine residues (Cys118, Cys126, Cys82’) that comprise the metal binding sites, which are located at each end of coiled coil.
The action of MerR at MerOP region
MerR make a bound with the DNA and RNA polymerase, is recruited but not able to form an open complex. There will be a conformational change induced by Hg(II), this will bind the ternary complex and will initiate the transcription of the genes.
In the absence of Hg(II) and MerR, the RNA polymerase will transcribe from the merR promoter, and this will increase the concentration of MerR in the cell. When MerR finally binds to the MerOP region, the transcription of the merR promoter is going to be repressed and consequently the DNA will bent and unfold at the MerOP sequence. The RNA polymerase will attach to the mer promoter, forming a complex between the DNA, MerR and RNA polymerase.
In the absence of Hg(II) and presence of MerR, this protein is going to bound the DNA in the repressor conformation, maintaining the repression of the promoter. The Hg(II) will bind to one of the two binding sites on the MerR, causing a conformational change to put MerR in the activation conformation. This will cause the DNA distortion at the centre of the operator, unwinding of DNA and straightening of the helix. The reorientation of the sequences so caused, allows them to interact with the RNA polymerase and initiate the transcription.
Modeling
Equations for de-greening regulatorsystem
An inducible two way promoter is connected downstream to a de-greening system and upstream to a regulator from the mer operon. This regulator, called merR, lead the expression of the PmerOT promoter.
In normal conditions, PmerOT promoter is induced by mercury in the cell and transcribes downstream. However, when HG is absent, PmerOT transcribes upstream and express the protein merR, which also works as an activator-repressor for PmerOT.
PmerOT repressed the expression upstream in a negative feedback. Reaction coul be expressed as:
Where, merR is in charge of inhibiting the transcription rate.
The negative feedback reaction can be expressed as:
Being f (merR), the predictive function for the transcription rate. From the Hill’s function, it can be inferred for a repressed promoter that:
Then, the function for merR messenger is given by:
Thus, the function behavior will be cyclic, autoregulating itself.
Equations for the downstream de-greening system
Downstream, PmerOT promoter will be activated by metallic mercury. Thus, the activity for the promoter downstream is given by the Hill’s function:
Where DPC, represents the De-greening protein complex. Starting from this, we can infer the concentration of the construct by the equations:
Chalase:
RCCR:
PAO:
And for GUN4:
 "
"