Team:Heidelberg/pages/Human Practice/igemathome
From 2014.igem.org
(→Evaluation of iGEM@home, a new tool in Science Communication) |
(→3. Direct feedback and quotes) |
||
| (24 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
=Abstract= | =Abstract= | ||
| - | iGEM@home is a distributed computing platform build upon the BOINC framework. Volunteers download a client software that receives and runs work packages distributed from a central server. Combining the computational capacity of at least hundreds of personal computers, this approach allows (1) future iGEM teams to run heavy computations without the need of access to a big server cluster and (2) offers the opportunity to spread information about Synthetic Biology and iGEM events and projects. By actively promoting the platform and constantly motivating a growing community of iGEM@home supporters, we managed to accumulated | + | iGEM@home is a distributed computing [http://www.igemathome.org/ platform] build upon the BOINC framework. Volunteers download a client software that receives and runs work packages distributed from a central server. Combining the computational capacity of at least hundreds of personal computers, this approach allows (1) future iGEM teams to run heavy computations without the need of access to a big server cluster and (2) offers the opportunity to spread information about Synthetic Biology and iGEM events and projects. By actively promoting the platform and constantly motivating a growing community of iGEM@home supporters, we managed to accumulated more than 1.5 TeraFLOPS of computational power and displayed about 40,000 screensaver slides to more than 400 community members. We conducted a survey for evaluation and found our project content well received by the community: 80% of first time BOINC users was also interested in the information on synthetic biology and over 60% of the users had never heard of iGEM before and got involved into synthetic biology via iGEM@home. The succes of the project was reflected and stimulated by the national and international press. |
| - | ''If you want to know more about the software that we developed please visit our Software Overview page, if you are rather interested in the modelling that we ran on iGEM@home and how it relates to our project, please visit the [[Team:Heidelberg/Modeling/Linker_Modeling|Linker modeling page]].'' | + | ''If you want to know more about the software that we developed please visit our [[Team:Heidelberg/Software/igemathome|Software Overview page]], if you are rather interested in the modelling that we ran on iGEM@home and how it relates to our project, please visit the [[Team:Heidelberg/Modeling/Linker_Modeling|Linker modeling page]]. If you want to become part of the iGEM@home community please visit [http://www.igemathome.org/ www.igemathome.org].'' |
| + | |||
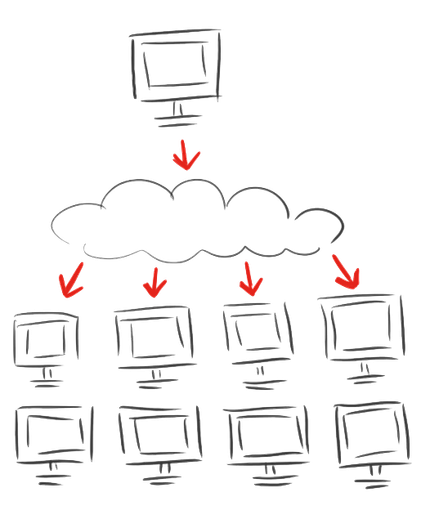
| + | {{:Team:Heidelberg/templates/image-quarter|align=right|caption=Principle of Distributed Computing|descr=|file=distributing_cloud_igemathome.org.png}} | ||
=Introduction= | =Introduction= | ||
| - | The creative concepts developed and the plethora of information gathered during the last decade by iGEM teams from all over the world proof efforts in Human Practices worth to be rewarded with a track of its own | + | The creative concepts developed and the plethora of information gathered during the last decade by iGEM teams from all over the world proof efforts in Human Practices worth to be rewarded with a track of its [https://2014.igem.org/Tracks/Policy_Practices own] in this year’s iGEM competition. Since iGEM teams from Heidelberg have always been interested in the question how laymen citizens can get involved into synthetic biology, they already introduced new concepts to the community to improve communication (first abstract page in multiple languages, first site tour for laymen, first mascot 2008), to do research on the mechanisms of science communication (2010) and to encourage reflection by philosophers and artists as representers of society (2010, 2013). Following the findings of our [https://static.igem.org/mediawiki/2010/7/72/Akzeptanz_end.pdf former team mates] |
, also supported by literature as described [http://www.sciencedirect.com/science/article/pii/S0016328713000384 here], that acceptance towards a technology is not correlated with the information provided to a sceptic public, we rather aim at involving the public into the creative process of scientific work. In the style of the new iGEM community labs track that encourages science amateurs to contribute “beyond the accolades of scientific publishing and economic reward”, we sought for a new way to involve laymen in actual science and build a strong community of well informed supporters and communicators of synthetic biology at the same time. Thereby we want to lay the foundation for a democratization of Science in an enlightened society. Now we proudly present the crowdsourcing and communication platform iGEM@home! | , also supported by literature as described [http://www.sciencedirect.com/science/article/pii/S0016328713000384 here], that acceptance towards a technology is not correlated with the information provided to a sceptic public, we rather aim at involving the public into the creative process of scientific work. In the style of the new iGEM community labs track that encourages science amateurs to contribute “beyond the accolades of scientific publishing and economic reward”, we sought for a new way to involve laymen in actual science and build a strong community of well informed supporters and communicators of synthetic biology at the same time. Thereby we want to lay the foundation for a democratization of Science in an enlightened society. Now we proudly present the crowdsourcing and communication platform iGEM@home! | ||
===Introducing Distributed Computing to the iGEM community=== | ===Introducing Distributed Computing to the iGEM community=== | ||
| - | Modern biology uses computational simulations of models for validation as well as to predict key experiments and suitable designs for specific applications. Despite of the fact that the experience with and the mere amount of algorithms and code dedicated to biological systems is growing exponentially these days, big data evaluation such as sequence analysis and multidimensional problems such as protein folding still consume vast amounts of computational resources. Therefore main aspects of biological science are only accessible to research institutions that provide these resources in form of supercomputers or computer clusters. In order to open computational biology up to all future iGEM teams in a manner that is not dependant of institutional affiliations, we decided to develop a platform from a set of applications that build upon the BOINC framework for distributed computing | + | Modern biology uses computational simulations of models for validation as well as to predict key experiments and suitable designs for specific applications. Despite of the fact that the experience with and the mere amount of algorithms and code dedicated to biological systems is growing exponentially these days, big data evaluation such as sequence analysis and multidimensional problems such as protein folding still consume vast amounts of computational resources. Therefore main aspects of biological science are only accessible to research institutions that provide these resources in form of supercomputers or computer clusters. In order to open computational biology up to all future iGEM teams in a manner that is not dependant of institutional affiliations, we decided to develop a platform from a set of applications that build upon the BOINC framework for distributed computing ([[Team:Heidelberg/Software/igemathome| Details on Software]]). Thereby we provide access to computing power for future (community lab) [http://www.igemathome.org/collaboration iGEM teams] and at the same time create an opening for laymen to jump in and empower science! |
===BOINC and iGEM - Foundations of a rewarding relationship=== | ===BOINC and iGEM - Foundations of a rewarding relationship=== | ||
| - | The BOINC framework distributes computational tasks to volunteers who provide computational resources in form of calculation time spent by the CPU of their idling personal computers. Volunteers download and install a client software that receives work packages from a central server unit and runs calculations once the computer is not in use. Alternatively users may dedicate a certain percentage of their computing power to destributed computing projects that they like to support. After the successful execution of the computation the result is uploaded to our server. One of the best known distributed computing projects is [http://setiathome.ssl.berkeley.edu/ SETI@home], a project in which volunteers contribute computing power to the search for extra-terrestrial intellegance. In our case the need for computing power arose from the calculations needed to evaluate the linker model that provides rigid linkers for the circularization of proteins | + | The BOINC framework distributes computational tasks to volunteers who provide computational resources in form of calculation time spent by the CPU of their idling personal computers. Volunteers download and install a client software that receives work packages from a central server unit and runs calculations once the computer is not in use. Alternatively users may dedicate a certain percentage of their computing power to destributed computing projects that they like to support. After the successful execution of the computation the result is uploaded to our server. |
| + | |||
| + | One of the best known distributed computing projects is [http://setiathome.ssl.berkeley.edu/ SETI@home], a project in which volunteers contribute computing power to the search for extra-terrestrial intellegance. In our case the need for computing power arose from the calculations needed to evaluate the [[Team:Heidelberg/Software/Linker_Software|linker model]] that provides rigid linkers for the circularization of proteins. In both cases the contributors are rewarded with points that reflect the computing time already spent and the dedication towards certain projects. In order to take motivation to the next level we implemented a platform that allows to feed information to the BOINC screensaver (figure 1). Thereby we enable iGEM teams from all over the world to reach an interested and motivated audience with information about their projects, Human Practices activities and iGEM and Synthetic Biology in general. In return users of iGEM@home will be rewarded with informations and insights into one of the most vital communities in science that catch their attention in the very moment they get back to their computers. | ||
===Outreach=== | ===Outreach=== | ||
| Line 103: | Line 107: | ||
{{:Team:Heidelberg/templates/image-quarter|align=right|caption=www.igemathome.org|descr=The <a href="http://www.igemathome.org/">homepage</a> of iGEM@home|file=igemathome_website_screenshot.PNG}} | {{:Team:Heidelberg/templates/image-quarter|align=right|caption=www.igemathome.org|descr=The <a href="http://www.igemathome.org/">homepage</a> of iGEM@home|file=igemathome_website_screenshot.PNG}} | ||
===Website=== | ===Website=== | ||
| - | A central part of was our campaign for iGEM@home is the website [http://www.igemathome.org www.igemathome.org] that is the frontend of our BOINC server and therefore needed to be externally hosted. (To find out more about the software click here). It bundles all important information on the [http://www.igemathome.org/project project] and explains iGEM in a layman’s way to an idea in what context this project was developed. It also provides a list of [http://www.igemathome.org/FAQ FAQ’s] that address most important concerns and questions that people have. In addition we also provide a list of [http://www.igemathome.org/related_projects other BOINC projects], because the client makes it very easy to support additional projects from different areas of science. Furthermore the website also hosts the [http://www.igemathome.org/igemathome/ “BOINC pages”]. These are the pages, where participants can view the server status, manage their account, create a profile and join the discussion in the [http://www.igemathome.org/igemathome/forum_index.php forums]. (For more information on the role of the forum in the outreach you can skip to [[#Forums]].) | + | A central part of was our campaign for iGEM@home is the website [http://www.igemathome.org www.igemathome.org] that is the frontend of our BOINC server and therefore needed to be externally hosted. (To find out more about the software click here). It bundles all important information on the [http://www.igemathome.org/project project] and explains iGEM in a layman’s way to an idea in what context this project was developed. It also provides a list of [http://www.igemathome.org/FAQ FAQ’s] that address most important concerns and questions that people have. In addition we also provide a list of [http://www.igemathome.org/related_projects other BOINC projects], because the client makes it very easy to support additional projects from different areas of science. Furthermore the website also hosts the [http://www.igemathome.org/igemathome/ “BOINC pages”]. These are the pages, where participants can view the server status, manage their account, create a profile and join the discussion in the [http://www.igemathome.org/igemathome/forum_index.php forums]. (For more information on the role of the forum in the outreach you can skip to [[#Forum:_Feedback_on_Computing_and_Projects|Forums]].) |
===Video=== | ===Video=== | ||
| Line 234: | Line 238: | ||
===2. Online survey on iGEM@home=== | ===2. Online survey on iGEM@home=== | ||
| - | Close to wiki freeze we | + | Close to wiki freeze we conducted an online survey to gather some additional information about the users and evaluate key features of iGEM at home. The majority of survey participants are male (88%) whilst they spread widely across the age spectrum (12-88 years old). 89% of participants decided to join the iGEM@home project to support the science of the iGEM teams. Only a few users are interested in the teamwork aspect of distributed computing or in comparing computer power with other users. The survey was well received by the community and covered 20% of the total users of iGEM@home, starting with 55 replies at the first day and dropping to less than 5 replys/day at the second day. |
| - | A set of results regarding the content and setup of iGEM at home is visualized in figure 1. The data in (1) reveals that most of the participants are users we want to call "BOINC veterans" that already run or ran several BOINC projects on their computers (82%). Comparison of the server data with the data automatically gathered by the iGEM@home user platform reveals that the group of BOINC | + | A set of results regarding the content and setup of iGEM at home is visualized in figure 1. The data in (1) reveals that most of the participants are users we want to call "BOINC veterans" that already run or ran several BOINC projects on their computers (82%). Comparison of the server data with the data automatically gathered by the iGEM@home user platform reveals that the group of BOINC veterans is slightly overrepresented in the survey (82% survey/75% total users). The two populations of users differ in their attitude towards the two communication platforms screensaver and forum which is represented in the bar chart (Figure 2). BOINC veterans equally pay attention to both media whilst new users tend to focus on the screensaver. The fact that 80% of new users reviewed the slide show is an encouraging result regarding the effectivity of content distribution via the screensaver (also compare figure 1,(2) and (3)). |
{{:Team:Heidelberg/templates/image-full|align=left|caption=Figure 1, Pie Charts (1-6): |descr=Pie charts that display data chosen from our online survey with a total set of 14 items and a total number of 82 (20%) participants from a total of 412 iGEM@home users. (1) More than 80% of users that filled out the survey were active members of the distributed computing community before joining iGEM@home. (2) Only 40% of the users paid active attention to the contents of the screensaver. (Please also see the bar chart below for more information). Still 70% of these users evaluated the screensaver as good or better. (3) The forum was evaluated by more than 55 % of the users of whose 60% gave positive reviews. (4) The computing tasks themselves (calculations) were positively rated by 65 % of the participants. Users point out a special need for improvements (in short: ‘work on!’) in the calculations (see comments in the text). (5) An overwhelming majority of about 80% of the participants liked the overall project and ranked it ‘good’ or even ‘perfect’ while none of the participant ranked the overall project as ‘bad’. (6) iGEM@home reached at least 52 persons that had no knowledge of iGEM before which is more than 60% of survey participants.|file=iah_hp_piecharts.JPG}} | {{:Team:Heidelberg/templates/image-full|align=left|caption=Figure 1, Pie Charts (1-6): |descr=Pie charts that display data chosen from our online survey with a total set of 14 items and a total number of 82 (20%) participants from a total of 412 iGEM@home users. (1) More than 80% of users that filled out the survey were active members of the distributed computing community before joining iGEM@home. (2) Only 40% of the users paid active attention to the contents of the screensaver. (Please also see the bar chart below for more information). Still 70% of these users evaluated the screensaver as good or better. (3) The forum was evaluated by more than 55 % of the users of whose 60% gave positive reviews. (4) The computing tasks themselves (calculations) were positively rated by 65 % of the participants. Users point out a special need for improvements (in short: ‘work on!’) in the calculations (see comments in the text). (5) An overwhelming majority of about 80% of the participants liked the overall project and ranked it ‘good’ or even ‘perfect’ while none of the participant ranked the overall project as ‘bad’. (6) iGEM@home reached at least 52 persons that had no knowledge of iGEM before which is more than 60% of survey participants.|file=iah_hp_piecharts.JPG}} | ||
| - | The review of the computational tasks/calculations in correspondence with direct feedback messages and posts of users via the comments field of the survey and the forum pages(see paragraph 3.) reveals a need for improvement of calculations concerning the way calculation processes are split into consumable subtasks. BOINC veterans | + | The review of the computational tasks/calculations in correspondence with direct feedback messages and posts of users via the comments field of the survey and the forum pages (see paragraph 3.) reveals a need for improvement of calculations concerning the way calculation processes are split into consumable subtasks. BOINC veterans reviewed the calculations (figure 1, (4)) as to be improved by introducing checkpoints to the calculations that allow an abandoned task to be continued instead of being recalculated from the very start. The overall reception of the project is very good. Not a single participant rated the project 'bad'. Only 5% of the participants did not know how to rank the BOINC project iGEM@home (figure 1, (5). One of the most imporatant results of the survey is the fact that more than 60% of the participants did not now of the existence of the iGEM competition before they downloaded and supported iGEM@home. |
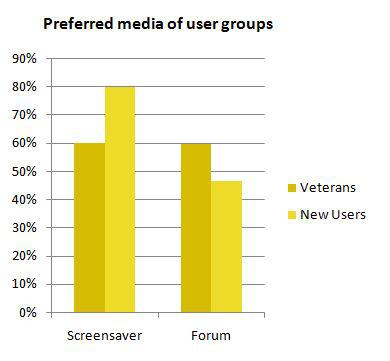
| + | {{:Team:Heidelberg/templates/image-quarter|align=right|caption=Figure 2, Bar Chart: |descr=The bar chart displays the numbers of participants that did evaluate the respective media (screensaver and forum). Whilst both media were evaluated by about 60% of BOINC ‘veterans’, new users preferably reviewed the screensaver (80%) and about 55% did not rank the forum at all.|file=iah_hp_vet_vs_new.JPG}} | ||
| + | ===3. Direct feedback and quotes=== | ||
| - | + | ''Comments and feedback on software and infrastructure:'' | |
| - | = | + | '''"Need checkpoints!":''' This statement was supported by additional 7 participants of the survey and also a major issue for [http://www.igemathome.org/igemathome/forum_thread.php?id=9 discussion] on the forum. Checkpoints are a feature strongly demanded by many BOINC veterans which is also represented in the review of the calculations (figure 1, (4) and will be implemented as soon as possible. |
| - | + | ||
| + | '''"Don't want or need a screensaver.":''' A statement supported by 2 users in total that are both supporters of additional BOINC projects. Some BOINC-enthusiasts that focus on collecting BOINC points might not be interested in the information spread via the screensaver. | ||
| + | |||
| + | '''"Forum spam was a VERY noticeable problem at first, but since the changes have been implemented as requested by various users I have not seen any spam in the forums.":''' It is important to quickly respond to requests by members of the community. The BOINC community is very communicative and eager to solve problems in coordination with developers. Several measures including the requirement for posters to have at least earned a few BOINC points with calculations, detecting and deleting spammers and active moderation of the forum solved the problem of spam effectively. | ||
| + | |||
| + | '''"Credit rate is poor compared to many other projects."''' Three users supported the claim that the points earned for computations for the iGEM@home project are low in comparison to other projects. The users proposed a dynamic point and award system (badges) for long-term motivation of BOINC crunchers. | ||
| + | |||
| + | '''"You should ensure that the server does not run out of tasks very often."''' Many users directly contacted us and asked for more tasks to calculate and commented that BOINC projects should not run out of tasks easily. We hope that due to the introduction of iGEM@home to the community, the servers will not run out of tasks easily during the summer. | ||
| + | |||
| + | ''Communication with the team and the overall organisation of the project was praised by many users:'' | ||
| + | |||
| + | '''"Great Project. Of course needs some time to get running, but definitely something worth supporting!"''' | ||
| + | |||
| + | '''"Very well done for a first BOINC project."''' | ||
| + | |||
| + | '''"I wish every project were this organized & committed."''' | ||
| + | |||
| + | "Feedback from the project administrators is excellent! Keep up the great work!!!!!" | ||
| + | |||
| + | ''Comments about iGEM and distributed computing:'' | ||
| + | |||
| + | '''"We need more distributed computing projects in biology and medical fields.":''' | ||
| + | |||
| + | '''"I did not know about synthetic biology until I participated on iGEM@home. Synthetic biology has great potential to benefit humanity that most people do not realize. Many people are still not aware of this new project. Time to spread the word out!"''' | ||
| - | = | + | =Conclusions= |
| - | We consider iGEM@home a great success. Not just were we able to do calculations impossible without a huge server cluster, we also reached a lot of people who didn’t know about Synthetic Biology and iGEM before. | + | We consider iGEM@home a great success. The elaborate evalution of the project showed that the product iGEM@home fulfiils all the requirements for a vivid distributed computing AND science communication plattform at the same time. Not just were we able to do calculations impossible without a huge server cluster, we also reached a lot of people who didn’t know about Synthetic Biology and iGEM before. At the moment the majority of iGEM@home users are experienced BOINC supporters and crunchers that also support additional projects. But we did not only show that we were able to recruit about 100 new BOINC supporters in a short time but that the content distributed via the screen saver slide show is accessed by many users and especially by the ones recruited directly for iGEM@home (80%). |
We hereby have shown that it is feasible for an iGEM team to run an distributed computing project and it offers a great new approach to reach out to the society. | We hereby have shown that it is feasible for an iGEM team to run an distributed computing project and it offers a great new approach to reach out to the society. | ||
Latest revision as of 01:58, 18 October 2014
Contents |
Abstract
iGEM@home is a distributed computing [http://www.igemathome.org/ platform] build upon the BOINC framework. Volunteers download a client software that receives and runs work packages distributed from a central server. Combining the computational capacity of at least hundreds of personal computers, this approach allows (1) future iGEM teams to run heavy computations without the need of access to a big server cluster and (2) offers the opportunity to spread information about Synthetic Biology and iGEM events and projects. By actively promoting the platform and constantly motivating a growing community of iGEM@home supporters, we managed to accumulated more than 1.5 TeraFLOPS of computational power and displayed about 40,000 screensaver slides to more than 400 community members. We conducted a survey for evaluation and found our project content well received by the community: 80% of first time BOINC users was also interested in the information on synthetic biology and over 60% of the users had never heard of iGEM before and got involved into synthetic biology via iGEM@home. The succes of the project was reflected and stimulated by the national and international press.
If you want to know more about the software that we developed please visit our Software Overview page, if you are rather interested in the modelling that we ran on iGEM@home and how it relates to our project, please visit the Linker modeling page. If you want to become part of the iGEM@home community please visit [http://www.igemathome.org/ www.igemathome.org].
Introduction
The creative concepts developed and the plethora of information gathered during the last decade by iGEM teams from all over the world proof efforts in Human Practices worth to be rewarded with a track of its own in this year’s iGEM competition. Since iGEM teams from Heidelberg have always been interested in the question how laymen citizens can get involved into synthetic biology, they already introduced new concepts to the community to improve communication (first abstract page in multiple languages, first site tour for laymen, first mascot 2008), to do research on the mechanisms of science communication (2010) and to encourage reflection by philosophers and artists as representers of society (2010, 2013). Following the findings of our former team mates , also supported by literature as described [http://www.sciencedirect.com/science/article/pii/S0016328713000384 here], that acceptance towards a technology is not correlated with the information provided to a sceptic public, we rather aim at involving the public into the creative process of scientific work. In the style of the new iGEM community labs track that encourages science amateurs to contribute “beyond the accolades of scientific publishing and economic reward”, we sought for a new way to involve laymen in actual science and build a strong community of well informed supporters and communicators of synthetic biology at the same time. Thereby we want to lay the foundation for a democratization of Science in an enlightened society. Now we proudly present the crowdsourcing and communication platform iGEM@home!
Introducing Distributed Computing to the iGEM community
Modern biology uses computational simulations of models for validation as well as to predict key experiments and suitable designs for specific applications. Despite of the fact that the experience with and the mere amount of algorithms and code dedicated to biological systems is growing exponentially these days, big data evaluation such as sequence analysis and multidimensional problems such as protein folding still consume vast amounts of computational resources. Therefore main aspects of biological science are only accessible to research institutions that provide these resources in form of supercomputers or computer clusters. In order to open computational biology up to all future iGEM teams in a manner that is not dependant of institutional affiliations, we decided to develop a platform from a set of applications that build upon the BOINC framework for distributed computing ( Details on Software). Thereby we provide access to computing power for future (community lab) [http://www.igemathome.org/collaboration iGEM teams] and at the same time create an opening for laymen to jump in and empower science!
BOINC and iGEM - Foundations of a rewarding relationship
The BOINC framework distributes computational tasks to volunteers who provide computational resources in form of calculation time spent by the CPU of their idling personal computers. Volunteers download and install a client software that receives work packages from a central server unit and runs calculations once the computer is not in use. Alternatively users may dedicate a certain percentage of their computing power to destributed computing projects that they like to support. After the successful execution of the computation the result is uploaded to our server.
One of the best known distributed computing projects is [http://setiathome.ssl.berkeley.edu/ SETI@home], a project in which volunteers contribute computing power to the search for extra-terrestrial intellegance. In our case the need for computing power arose from the calculations needed to evaluate the linker model that provides rigid linkers for the circularization of proteins. In both cases the contributors are rewarded with points that reflect the computing time already spent and the dedication towards certain projects. In order to take motivation to the next level we implemented a platform that allows to feed information to the BOINC screensaver (figure 1). Thereby we enable iGEM teams from all over the world to reach an interested and motivated audience with information about their projects, Human Practices activities and iGEM and Synthetic Biology in general. In return users of iGEM@home will be rewarded with informations and insights into one of the most vital communities in science that catch their attention in the very moment they get back to their computers.
Outreach
The need to build a vivid community is key to run a successful distributed computing project. As the project depends on the voluntary support of people it is important to get people enthused about the science behind the project, so they are willing to support the project. Good communication is therefore essential to find support. We therefore used different channels to present and promote our project and iGEM in general to convince people to help us - which in the end turned out to be key to one of our biological main projects!
The Communication Platform iGEM@HOME
The important job after convincing the user to try our software, is to convince them to continuously support us by constantly feeding them with interesting facts on iGEM and offering them ways to influence the project.
Forum: Feedback on Computing and Projects
The first pillar for the communication is an integrated [http://www.igemathome.org/igemathome/forum_index.php forum] software, that serves several purposes: First of all, additional information like status updates or information on the biological projects can be communicated to the community via the 'News' and 'Science' sections of the forum. In addition the users can give elaborate feedback on problems arising from the client site. Finally user forums are an integral component to the BOINC distributed computing community and are widely used to present user achievments such as the number of projects supported, computational time spent to projects, BOINC points rewarded for computing and also to compare the stats of different projects [http://boincstats.com/en/stats/projectStatsInfo projects]. Forum users are provided with a personal profile that displays memberships in BOINC teams, points earned for computing and the possibility to contact other users via private text messages.
Screen Saver: Communication Channel for Synthetic Biology
The second pillar of our communicational concept is a browser page that feeds content to the [http://www.igemathome.org/screensaver screensaver] implemented into BOINC. We decided to post different types of content: (1) General information about (synthetic) biology, (2) general facts about the iGEM competition, (3) presentations of iGEM team projects, (4) announcements of special events that are open to public organised by iGEM (Human Practices) teams. We started with a small series of 'Did you know ...?' pages, presented the project of iGEM team Heidelberg 2013 and contacted other iGEM teams to offer the opportunity to present content to a growing community of iGEM supporters. In order to test the capabilities of iGEM@home as a communication channel, we announced our own events via the channel. A final test will be carried out after wiki freeze by announcing an event exclusively via the screensaver (see a short description in the 'Outlook' section at the end of this text). Last not least we can announce that we received content from two cooperating iGEM teams that we proudly present in our screensaver: Team Marburg sent a flyer for their 'iGEM Quiz Marburg' and the Berlin team sent in a text and figures on their project on 'Magnetic E.coli'.
Some example pages of the iGEM@home screensaver:
Promoting iGEM@home: Material and Methods
The homepage of iGEM@home
Website
A central part of was our campaign for iGEM@home is the website [http://www.igemathome.org www.igemathome.org] that is the frontend of our BOINC server and therefore needed to be externally hosted. (To find out more about the software click here). It bundles all important information on the [http://www.igemathome.org/project project] and explains iGEM in a layman’s way to an idea in what context this project was developed. It also provides a list of [http://www.igemathome.org/FAQ FAQ’s] that address most important concerns and questions that people have. In addition we also provide a list of [http://www.igemathome.org/related_projects other BOINC projects], because the client makes it very easy to support additional projects from different areas of science. Furthermore the website also hosts the [http://www.igemathome.org/igemathome/ “BOINC pages”]. These are the pages, where participants can view the server status, manage their account, create a profile and join the discussion in the [http://www.igemathome.org/igemathome/forum_index.php forums]. (For more information on the role of the forum in the outreach you can skip to Forums.)
Video
The second pillar of our promotion is a video that provides the information in an easily digestible way.
The video presents iGEM, our project and distributed computing. It explains why we need the support of the users and how to help us.
Flyer
To promote iGEM@home on local level to friends, fellow students and people interested in science and iGEM we created Flyers that are supposed to make people interested in the project and lead them (via QR-code or link) to our website.




Media Campaign
For the well prepared initial kickoff we addressed mailing lists in the context of iGEM and BOINC as well as distributing information via the iGEM team Heidelberg 2014 Facebook and Twitter accounts and via partner communities. In addition, our project was covered by 2 journals ([http://www.spektrum.de/news/proteinforschung-zum-mitmachen/1307953 Spektrum der Wissenschaft] german popular science magazine and [http://www.biotechniques.com/news/Synthetic-Biology-You-Can-Contribute-Too/biotechniques-354291.html#.VC7gDedXgXk BioTechniques] an international journal on Life Science Methods).
Evaluation of iGEM@home, a new tool in Science Communication
In order to evaluate our Human Practices approach, we investigated the perception of iGEM@home in three ways:
- We monitored the activity of users
- We conducted an evaluative survey
- We screened the content of the forum for important feedback
Here we list the most relevant results of our evaluation.
1. Website and client activity monitoring
(as of 12.10.2014)
User: The iGEM@home website currently counts more than 400 users from 42 countries. About 75% of supporters do also support other iGEM Projects. The Users are organized in more then 80 teams that build smaller communities and associations inside the BOINC community. The Age ranges from 12 to 88 years.
Computation power: More than 1,100 calculating computers are connected to the iGEM@home network and more than 100,000 jobs have already been computed in this project consuming 1.2 Million CPU hours. This is equivalent to a 28,190$ per month cloud computer cluster by amazon web services
Communication: Users of iGEM@home already posted a 190 times on into the forum (mostly providing bug reports and suggestions for further development steps). The screensaver has shown up 1300 times for an average of 2:08 hours. In this time it displayed about 40,000 screensaver slides.
For the most up to date information visit the [http://www.igemathome.org/igemathome/server_status.php server status page], the official [http://boincstats.com/en/stats/155/project/detail/overview BOINCstats] and our [http://igemathome.org/igemathome/forum_index.php forum].
2. Online survey on iGEM@home
Close to wiki freeze we conducted an online survey to gather some additional information about the users and evaluate key features of iGEM at home. The majority of survey participants are male (88%) whilst they spread widely across the age spectrum (12-88 years old). 89% of participants decided to join the iGEM@home project to support the science of the iGEM teams. Only a few users are interested in the teamwork aspect of distributed computing or in comparing computer power with other users. The survey was well received by the community and covered 20% of the total users of iGEM@home, starting with 55 replies at the first day and dropping to less than 5 replys/day at the second day.
A set of results regarding the content and setup of iGEM at home is visualized in figure 1. The data in (1) reveals that most of the participants are users we want to call "BOINC veterans" that already run or ran several BOINC projects on their computers (82%). Comparison of the server data with the data automatically gathered by the iGEM@home user platform reveals that the group of BOINC veterans is slightly overrepresented in the survey (82% survey/75% total users). The two populations of users differ in their attitude towards the two communication platforms screensaver and forum which is represented in the bar chart (Figure 2). BOINC veterans equally pay attention to both media whilst new users tend to focus on the screensaver. The fact that 80% of new users reviewed the slide show is an encouraging result regarding the effectivity of content distribution via the screensaver (also compare figure 1,(2) and (3)).
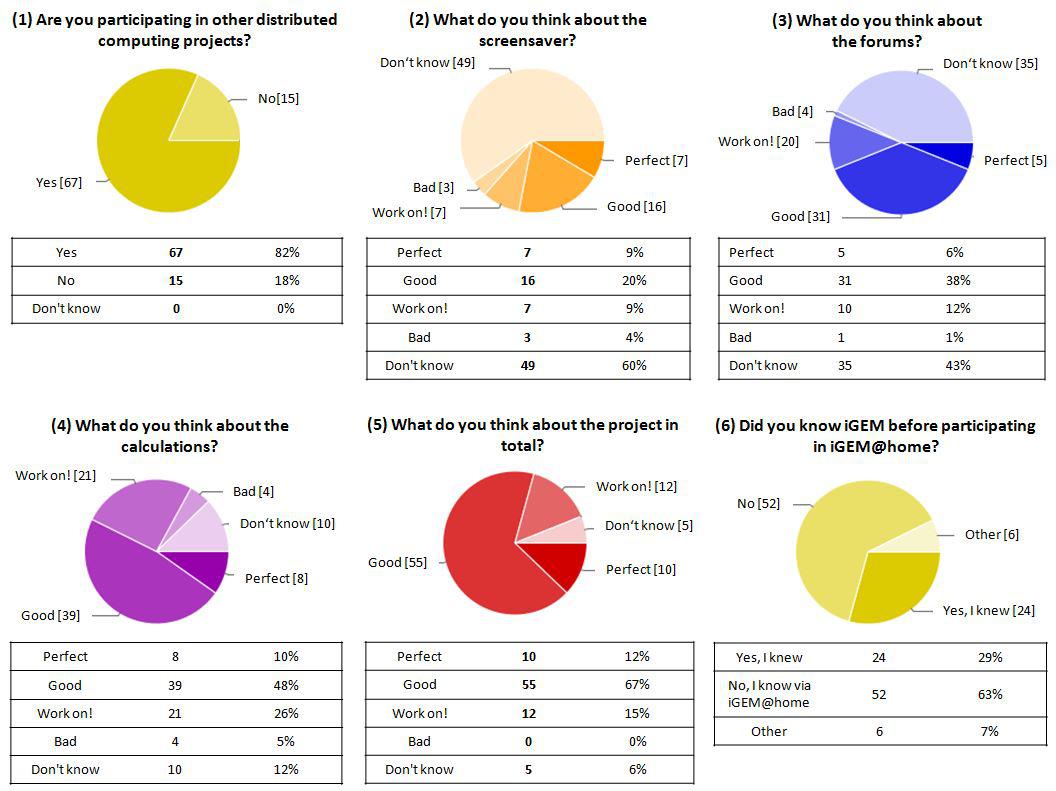
Pie charts that display data chosen from our online survey with a total set of 14 items and a total number of 82 (20%) participants from a total of 412 iGEM@home users. (1) More than 80% of users that filled out the survey were active members of the distributed computing community before joining iGEM@home. (2) Only 40% of the users paid active attention to the contents of the screensaver. (Please also see the bar chart below for more information). Still 70% of these users evaluated the screensaver as good or better. (3) The forum was evaluated by more than 55 % of the users of whose 60% gave positive reviews. (4) The computing tasks themselves (calculations) were positively rated by 65 % of the participants. Users point out a special need for improvements (in short: ‘work on!’) in the calculations (see comments in the text). (5) An overwhelming majority of about 80% of the participants liked the overall project and ranked it ‘good’ or even ‘perfect’ while none of the participant ranked the overall project as ‘bad’. (6) iGEM@home reached at least 52 persons that had no knowledge of iGEM before which is more than 60% of survey participants.
The review of the computational tasks/calculations in correspondence with direct feedback messages and posts of users via the comments field of the survey and the forum pages (see paragraph 3.) reveals a need for improvement of calculations concerning the way calculation processes are split into consumable subtasks. BOINC veterans reviewed the calculations (figure 1, (4)) as to be improved by introducing checkpoints to the calculations that allow an abandoned task to be continued instead of being recalculated from the very start. The overall reception of the project is very good. Not a single participant rated the project 'bad'. Only 5% of the participants did not know how to rank the BOINC project iGEM@home (figure 1, (5). One of the most imporatant results of the survey is the fact that more than 60% of the participants did not now of the existence of the iGEM competition before they downloaded and supported iGEM@home.
3. Direct feedback and quotes
Comments and feedback on software and infrastructure:
"Need checkpoints!": This statement was supported by additional 7 participants of the survey and also a major issue for [http://www.igemathome.org/igemathome/forum_thread.php?id=9 discussion] on the forum. Checkpoints are a feature strongly demanded by many BOINC veterans which is also represented in the review of the calculations (figure 1, (4) and will be implemented as soon as possible.
"Don't want or need a screensaver.": A statement supported by 2 users in total that are both supporters of additional BOINC projects. Some BOINC-enthusiasts that focus on collecting BOINC points might not be interested in the information spread via the screensaver.
"Forum spam was a VERY noticeable problem at first, but since the changes have been implemented as requested by various users I have not seen any spam in the forums.": It is important to quickly respond to requests by members of the community. The BOINC community is very communicative and eager to solve problems in coordination with developers. Several measures including the requirement for posters to have at least earned a few BOINC points with calculations, detecting and deleting spammers and active moderation of the forum solved the problem of spam effectively.
"Credit rate is poor compared to many other projects." Three users supported the claim that the points earned for computations for the iGEM@home project are low in comparison to other projects. The users proposed a dynamic point and award system (badges) for long-term motivation of BOINC crunchers.
"You should ensure that the server does not run out of tasks very often." Many users directly contacted us and asked for more tasks to calculate and commented that BOINC projects should not run out of tasks easily. We hope that due to the introduction of iGEM@home to the community, the servers will not run out of tasks easily during the summer.
Communication with the team and the overall organisation of the project was praised by many users:
"Great Project. Of course needs some time to get running, but definitely something worth supporting!"
"Very well done for a first BOINC project."
"I wish every project were this organized & committed."
"Feedback from the project administrators is excellent! Keep up the great work!!!!!"
Comments about iGEM and distributed computing:
"We need more distributed computing projects in biology and medical fields.":
"I did not know about synthetic biology until I participated on iGEM@home. Synthetic biology has great potential to benefit humanity that most people do not realize. Many people are still not aware of this new project. Time to spread the word out!"
Conclusions
We consider iGEM@home a great success. The elaborate evalution of the project showed that the product iGEM@home fulfiils all the requirements for a vivid distributed computing AND science communication plattform at the same time. Not just were we able to do calculations impossible without a huge server cluster, we also reached a lot of people who didn’t know about Synthetic Biology and iGEM before. At the moment the majority of iGEM@home users are experienced BOINC supporters and crunchers that also support additional projects. But we did not only show that we were able to recruit about 100 new BOINC supporters in a short time but that the content distributed via the screen saver slide show is accessed by many users and especially by the ones recruited directly for iGEM@home (80%).
We hereby have shown that it is feasible for an iGEM team to run an distributed computing project and it offers a great new approach to reach out to the society.
Future
Of course we think that iGEM@home is a great tool and needs to be continued after the end of this year’s iGEM competition. As it has an active and interested community and offers teams to do impressive computations, we hope to hand iGEM@home over to the headquarters for a centralized access in future. So that it becomes an integral part of iGEM, for the benefit of society.
 "
"