Team:HNU China/Project
From 2014.igem.org
(Difference between revisions)
Yannickchou (Talk | contribs) |
|||
| (45 intermediate revisions not shown) | |||
| Line 185: | Line 185: | ||
padding:0px; | padding:0px; | ||
} | } | ||
| + | .STYLE2 { | ||
| + | font-family: Times New Roman; | ||
| + | font-size: 17px; | ||
| + | } | ||
| + | .STYLE3 { | ||
| + | font-family: Arial, Helvetica, sans-serif; | ||
| + | font-size: 30px; | ||
| + | font-weight: bold; | ||
| + | font-style: italic; | ||
| + | } | ||
| + | .STYLE4 {color: #FF0000} | ||
</style> | </style> | ||
<script type="text/javascript" src="http://code.jquery.com/jquery-latest.js"></script> | <script type="text/javascript" src="http://code.jquery.com/jquery-latest.js"></script> | ||
| - | <script type="text/javascript" src=" | + | <script type="text/javascript" src="https://2014.igem.org/wiki/index.php?title=Team:HNU_China/ui.js&action=raw&ctype=text/javascript"></script> |
| - | <script type="text/javascript" src=" | + | <script type="text/javascript" src="https://2014.igem.org/wiki/index.php?title=Team:HNU_China/sortable.js&action=raw&ctype=text/javascript"></script> |
| - | <script type="text/javascript" src=" | + | <script type="text/javascript" src="https://2014.igem.org/wiki/index.php?title=Team:HNU_China/metadata.js&action=raw&ctype=text/javascript"></script> |
| - | <script type="text/javascript" src=" | + | <script type="text/javascript" src="https://2014.igem.org/wiki/index.php?title=Team:HNU_China/mbTabset.js&action=raw&ctype=text/javascript"></script> |
<script type="text/javascript"> | <script type="text/javascript"> | ||
$(function(){ | $(function(){ | ||
| Line 257: | Line 268: | ||
</div> | </div> | ||
<div class="wrapper"> | <div class="wrapper"> | ||
| - | <div class="tabset" id="tabset1"><a id="a" class="tab sel {content:'cont_1'}"> | + | <div class="tabset" id="tabset1"><a id="a" class="tab sel {content:'cont_1'}"><b>Background</b></a><a id="b" class="tab {content:'cont_2'}"><b>Description</b></a><a id="c" class="tab {content:'cont_3'}"><b>Experiment</b></a><a id="d" class="tab {content:'cont_4'}" ><b>Result</b></a><a id="a" class="tab {content:'cont_5'}" ><b>Future work</b></a></div> |
<div id="cont_1"> | <div id="cont_1"> | ||
| - | <div id="chead"></div> | + | <div id="chead"> |
| - | <div id="cmiddle"> | + | <div align="left"class="STYLE3" style="margin-left:50px;"><br/><br/><br/>Background</div> |
| + | </div> | ||
| + | <div id="cmiddle"> | ||
| + | <div align="left" class="STYLE2" style="margin-left:50px;margin-right:50px;"> | ||
| + | |||
| + | <b>Introduction</b><br><br> | ||
| + | |||
| + | Biomining, is used to describe the novel approach in mining industry when microorganisms are applied to the extraction and recovery of precious and base metals from ores and concentrates[fig.1]. Technicially, it consists two branches, the bioleaching and biooxidation. The bioleaching strictly refers to the case when microorganisms are used to solubilize the metal. While biooxidation mainly focus on the pretreatment of target metals by bio-processsing minerals that occlude target metals, such as thiosulfate encompasses the gold. <br><br> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2014/7/7d/HNUBackground1.jpg" alt="Background1" /><br> | ||
| + | Fig.1 Biomining is used to extract copper from copper ore.<br><br> | ||
| + | |||
| + | Although the same biological process had been unknowingly used to extract metals at mine sites in, for example Spain, the UK and China, for several hundred years, the modern era of bioming began with the discovery of the bactreium, <i>Thiobacillus ferrooxidans</i>(now <i>Acidithiobacillus ferrooxidans</i>) in the mid-1940s. In 1958 Kennecott Mining Company patented the use of <i>Thiobacillus ferrooxidans</i> for copper extraction from waste rock dumps at the Bingham Canyon mine in Utah, USA. A recent survey illustrates that currently bioming is commercially praticed in the production of 15% of copper, 5% of gold.<br><br> | ||
| + | |||
| + | <b>Why biomine?</b><br><br> | ||
| + | |||
| + | Bio-extractive techniques have to compete with alternative approaches for extracting metals from ores and concentrates. Some, such as pyrometallurgical technologies[fig.2] (ore roasting/smelting) have been refined over millennia and often represent major investments by mining companies, while others, such as pressure leaching, are more recent non-biological innovations. Main microorganisms involved in mineral oxidation processes are autotrophs, and the processes operate usually at atmospheric pressure and at relatively low temperatures (20–80 °C). Biomining is generally perceived as a much more environmentally benign (‘green’) approach, involving much lower temperatures (and hence energy costs) and smaller carbon footprints, which contrasts with current biomining operations, relied on the blasting and grinding of ore bodies, emitting large amounts of CO2, consuming 5% of total global energy production. Bio- processing also has niche advantages in two circumstances: first, when the ore or concentrate contains significant quantities of arsenic; second, for processing lowgrade and complex (polymetallic) ores.<br><br> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2014/d/d2/HNUBackground2.jpg" alt="Background2" /><br> | ||
| + | Fig2. Pyrometallurgy<br><br> | ||
| + | |||
| + | <b>Configurations and microbiology for biomining</b><br><br> | ||
| + | |||
| + | Engineering options for biomining have evolved from relatively inexpensive, partly controlled, irrigated dump or heap reactors to sophisticated, highly controlled and expensive stirred-tank reactors. Another distinguished way of bioming is the <i>in situ</i> mining, used extensively in Canada in the 1970s to recover uranium from worked out deep mines, which may well be the next major development in the mining sector. Mineral heaps and stirred tanks provide very different environments and challenges for mineral-leaching microorganisms, and different ‘optimal’ populations might be expected to emerge with similar target minerals depending on the reactor used. <br><br> | ||
| + | |||
| + | The extreme physico-chemical nature of bioleach liquors — low pH, elevated concentrations of (toxic) metals, metalloids and other solutes, and highly positive redox potential (EH values may exceed +900 mV) — means that they are highly toxic to the vast majority of life forms, including microorganisms. Microorganisms oxidize both sulfur and iron of sulfide minerals, such as pyrite. It is now well established that bioleaching and biooxidation in all biomining operations is mediated by consortia of acidophilic prokaryotes. These have been categorized as: firstly, ferric iron-generating autotrophs which produce the mineral oxidant; secondly, sulfuric acid-generating autotrophs, which maintain the low pH environment required; and finally, heterotrophic and mixotropic prokaryotes, which degrade organic compounds leaked from autotrophic iron-oxidizers and sulfur-oxidizers, there avoiding potential toxicity issues.<br><br> | ||
| + | |||
| + | <b>Engineered microbial consortia</b><br><br> | ||
| + | |||
| + | The field of synthetic biology has developed a wide range of highly engineered clonal populations of bacteria to perform complex tasks.The construction and analysis of synthetic gene circuits has not only provided us with new tools for genetic engineering but has given deeper insight into naturally occurring gene circuits, their evolution, architectures, and properties as well. The industrial practice of biomining and bioremediation of heavy metal contaminations could potentially benefit from synthetic consortia as natural consortia have been shown to play crucial roles in these processes. <br><br> | ||
| + | |||
| + | To our best knowledge, hybrid consortia consisting of genetically engineered and naturally occurring bioleaching bacteria have not been reported so far. In fact only two knockouts and two expression mutants have been reported in the scientific literature. One <i>rus</i> overexpressing <i>A. fer</i>rooxidans strain and another expressing the mer determinant for a mercury resistant A. caldus strain. Once more suitable transformation protocols have been developed, it maybe feasible to modulate quorum-sensing signals with engineered microbes by either attenuating or amplifying natural signals or sending artificial signals to promote biofilm formation or mobilization respectively as recently demonstrated with engineered <i>E.coli</i> cells.<br><br> | ||
| + | |||
| + | <b>Fungi in acid mine drainage</b><br><br> | ||
| + | |||
| + | Roles of fungi, the natural residents of acid mine drainage(AMD) and its attenuator are not emphasized adequately in the mine water research. Though AMD appears to be a certain special enviroment with relatively high metal concentration as well low pH, several species of fungi are also isolated from the AMD carrying streams, even AMD. Generally, fungi occur over a wide pH range (pH 1.0–11.0) and have been detected in acid habitats like volcanic springs, acid mine drainage or acid industrial wastewaters. Many of them are primarily acid-tolerant, but truly acidophilic species have also been detected[talbe.1]. While extracellular precipitation, complexation and crystallization, metal transformation, biosorption and sequestration are seen for fungi, endow fungi the ability to survive. <br><br> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2014/5/5f/HNUBackground3.jpg" alt="Background3" /><br><br> | ||
| + | |||
| + | Under this condition, fungi may play the important role as primary degraders of complex organic matter, due to the absence of invertebrates that actively shred the leaves (shredders) at pH values below 3.5. At the same time, the fungi will contribute to oxygen consumption, thereby limiting oxidative stress for the anaerobic bactiria. Moreover, fungi can be directly involved in the reduction of ferric iron or sulphur. Comparatively, more reports are available concerning the absorption of heavy metals by fungi, in comparison to bacteria or algae in freshwater ecosystem. Fungi can absorb metals in their cell wall or adsorb in extracellular polysaccharide slime. This capacity enables them to grow in the presence of high amounts of heavy metals. Fungal activity in acid mine drainage is represented in Fig.3<br><br> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2014/7/7c/HNUBackground4.jpg" alt="Background4" /><br> | ||
| + | Fig.3 Schematic diagram of fungal influence in acid mine drainage remediation<br><br> | ||
| + | |||
| + | <b>Conclusion</b><br><br> | ||
| + | |||
| + | Biomining, is used to describe the novel approach in mining industry when microorganisms are applied to the extraction and recovery of precious and base metals from ores and concentrates. Compared with the conventional approachs, such as pyrometallurgical technologies and pressure leaching in mining industry, bioming has its unique advantages in energy conservation and emission reduction, both of which are definitely with rising importance in the modern time. Microorganisms are usually settled in the non-sterile condition, and participate the bio-processing in consortia. In most of the time, they play the role of iron-oxidizer, sulfur-oxdizer and mixotrophic or heterotrophic acidophiles repectively or together. In the “bottom up” approach to optimize the consortia, the logic of synthetic biology is a worthy try to get the optimum colonies. Fungi is a natural resident in the acid mine drainage and involved in the reduction of ferric iron and sulphur, matching the main role in bioming consortia. Among these fungi is <i>Saccharomyces cerevisiae</i>, one of the best studied species and a very popular model organism for synthetic biology. The goal of our project is to take advantage of synthetic biology approach to synthesize a optimized biomining <i>Saccharomyces cerevisiae strain</i>.<br><br> | ||
| + | |||
| + | <b>Reference</b> </br> | ||
| + | <ol> | ||
| + | <li>Biomining — biotechnologies for extracting and recovering metals from ores and waste materials. D Barrie Johnson. Current Opinion in Biotechnology 2014, 30:24–31 </li> | ||
| + | <li>The microbiology of biomining: development and optimization of mineral-oxidizing microbial consortia. Douglas E. Rawlings1 and D. Barrie Johnson2.Microbiology (2007), 153, 315–324 </li> | ||
| + | <li>How will biomining be applied in future? C. L. BRIERLEY. Trans. Nonferrous Met. Soc. China 18(2008) 1302-1310 </li> | ||
| + | <li>Engineering microbial consortia to enhance biomining and bioremediation. Karl D. Brune andTravis S. Bayer* . Frontiers in Microbiology. 05 June 2012</li> | ||
| + | <li>Occurrence and role of algae and fungi in acid mine drainage environment with special reference to metals and sulfate immobilization. Bidus Kanti Dasa, Arup Roya, Matthias Koschorreckb, Santi M. Mandalc, Katrin Wendt-Potthoffb, Jayanta Bhattacharyaa,*. water research 43 (2009)883–894</li> | ||
| + | </ol> | ||
| + | </br> | ||
| + | </div> | ||
| + | </div> | ||
<div id="cfoot"></div> | <div id="cfoot"></div> | ||
</div> | </div> | ||
| + | |||
<div id="cont_2"> | <div id="cont_2"> | ||
| - | <div id="chead"></div> | + | <div id="chead"> |
| - | <div id="cmiddle">2</div> | + | <div align="left"class="STYLE3" style="margin-left:50px;"><br/><br/><br/>Description</div> |
| + | </div> | ||
| + | <div id="cmiddle"> | ||
| + | <div align="left" class="STYLE2" style="margin-left:50px;margin-right:50px;"> | ||
| + | |||
| + | <b>I.Overview</b><br><br> | ||
| + | |||
| + | Biomining refers to new technologies that microorganisms are applied to extract and recover metals from ores and concentrates. In mordant mining industry, the application of biomining is rapidly increased, since it has the advantage of energy conservation and emission reduction. In biomining, microorganisms are mainly used as iron-oxidizer or sulfur-oxidizer to solubilize either target metals or the unwanted minerals that occlude target metals. Microorganisms has also been applied to recover metal from the abandoned ore or slags, and remediate Acid Mine Drainage (AMD). Fungi are the natural residents of AMD. With the ability of ferric iron oxidation and absorption, they can also take part into the biomining process and the following metal reduction operation. Saccharomyces cerevisiae is one of the fungi species that has been identified in the AMD, yet its utility in biomining has largely been ignored. Here, we genetically engineer <i>Saccharomyces cerevisiae</i> to enhance its capacity of ferric iron oxidation and absorption. Meanwhile, we also introduce an inducible apoptosis system into the yeast so that unwanted or escape transgenic yeasts can be easily distroyed. Traditional method used to optimize microbial consortium for biomining is to inculate indigenous microflora into the reactor and screen for the better recombination. Compared with this so call “top down” method, our approach takes advantage of the powerful yeast genetic engineering to logically and systemically improve the desired traits. <br><br> | ||
| + | |||
| + | We name our designed Saccharomyces cerevisiae strain ‘microbial miner’. This microbial miner consists of two parallel systems, iron sensitive absorbing system and optogenetic apoptosis system.<br><br> | ||
| + | |||
| + | <b>II.Iron sensitive aborbing system</b><br><br> | ||
| + | |||
| + | <b>1.FET3 protein</b><br><br> | ||
| + | |||
| + | High affinity iron uptake in <i>Saccharomyces cerevisiae</i> requires FET3 protein. Localized on the external cell surface, FET3 protein is proposed to facilitate iron uptake by catalyzing the oxidation of Fe(II) to Fe(III) by O2, shown as below:<br><br> | ||
| + | |||
| + | 4Fe(II) + O2 + 4H+ → 4Fe(III) + 2H2O<br><br> | ||
| + | |||
| + | The posttranslational insertion of four copper ions into FET3 protein is essential for its activity, thus linking copper and iron homeostasis together[fig.1]. In this model, Fe(III) is the substrate for the iron permease, encoded by FTR1 protein[fig2]. <br><br> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2014/8/82/HNUDiscription1.jpg" alt="Discription1"/> <br> | ||
| + | Fig.1 Ribbon diagram of FET3 protein. The T1, T2, and T3 copper atoms are shown in blue, green, and yellow, respectively. The three plastocyanin-like domains are shown as blue, green, and gold. Included in red are the carbohydrates resolvable in the electron density map.<br><br> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2014/3/38/HNUDiscription2.jpg" alt="Discription2"/> <br> | ||
| + | Fig.2 FET3 and FTR1 proteins assemble into a complex in a cellular compartment early in the secretory pathway, most likely in the ER. The complex progresses to a post-Golgi compartment, where the CCC2 protein mediates copper delivery to FET3 protein. Finally, the copper-loaded F ET3 protein, presumably still complexed to FTR1 protein, is delivered to the plasma membrane and becomes competent for iron transport. <br><br> | ||
| + | |||
| + | <b>2.IRE-IRP1</b><br><br> | ||
| + | |||
| + | <i>Saccharomyces cerevisiae</i> post-translationally regulates the expression of the plasma membrane high affinity iron transport system, consisting FET3 protein and FTR1 protein, in response to iron need. Incubation of cells in high iron leads to the internalization and degradation of both FET3 protein and FTR1 protein. Since we are looking for a way to accumulate iron within the cell, the cell should promote the absorption with the presence of iron. We thus cast our eyes to other mammalian iron regulation system. <br><br> | ||
| + | |||
| + | In human cell, the cellular levels of ferritin and transferrin receptor (TfR) are also primarily regulated at translational level upon changes in iron availability, through interactions between iron regulatory proteins (IRP1 and IRP2) and iron responsive elements (IRE) in the 5’ UTR of H- and L-ferritin mRNA and the 3’ UTR of TfR mRNA. When intracellular iron levels rise, IRP1 can’t bind IRE, IRP2 is degraded and ferritin mRNA is efficiently translated. [fig.3] When iron level is high, IRP1 exists as a cytosolic aconitase. When iron is low, it assumes an open configuration associated with the loss of iron atoms in the iron-sulfur cluster, and can bind the IRE stem loop.[fig.4]<br><br> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2014/a/ac/HNUDiscription3.jpg" alt="Discription3"/> <br> | ||
| + | Fig.3 Decreased iron supply activates binding of IRPs to IRE resulting in translational inhibition of the mRNAs encoding ferritin and stabilization of the Tf R mRNA. During high iron conditions, IRPs lose their affinity for IREs, increasing translation of ferritin mRNAs and degradation of the Tf R mRNA.<br><br> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2014/e/eb/HNUDiscription4.jpg" alt="Discription4"/> <br> | ||
| + | Fig.4 The Fe-S cluster in aconitase.<br><br> | ||
| + | |||
| + | <b>3.Our project</b><br><br> | ||
| + | |||
| + | We plan to introduce IRE-IRP1 system into <i>Saccharomyces cerevisiae</i> to control the translation of FET3 mRNA, enhancing its iron aborption capability. In the circumstance of low iron concentration, the constitutively expressing IRP1 bonds to the stem loop of IRE preceding the FET3 mRNA, impeding the translation of FET3. Once iron reaches to a high level, the breakdown of IRE-IRP1 interaction shall activate the FET3 translation, which lead to more iron uptake, at last achieve the goal for iron accumulation.[fig.5]<br><br> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2014/0/0a/HNUDiscription5.jpg" alt="Discription5"/> <br> | ||
| + | Fig.5 Iron sensitive aborbing system<br><br> | ||
| + | |||
| + | <b>III.Optogenetic apoptosis system</b><br><br> | ||
| + | |||
| + | <b>1.Caspase-3</b><br><br> | ||
| + | |||
| + | Apoptosis or programmed cell death (PCD) is a highly coordinated cellular suicide program. Caspases, cysteine-dependent aspartate-directed proteases are a family of cysteine proteases that play essential roles in apoptosis[fig.6]. In previous studies in yeast, only the over-expression of caspase 8 led to cell death while the over-expression of caspase 3 was not lethal and only retarded yeast growth. In our design, the apoptosis related protein expresses when exposed to the blue light. Totally dark is harsh to fulfill in the industrial practicing, and urge us put the possibility of expression leakage into consideration, when the toxity of caspase may interfere the iron uptaking. Hence, we choose caspase-3, instead of caspase-8, as the apoptosis trigger. <br><br> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2014/2/27/HNUDiscription6.jpg" alt="Discription6"/> <br> | ||
| + | Fig.6 Extrinsic and intrinsic pathways to caspase-3 activation.<br><br> | ||
| + | |||
| + | Although yeasts lack some elements of the complex apoptotic machinery of metazoan cells, recent studies show that many features of apoptosis, including a caspase-like activity, can be induced in these organisms by DNA damage and other apoptotic triggers. Here we plan to introduce CASP3 gene from human, and attempt to verify our hypothesis of the apoptosis effect in yeast by Caspase-3.<br><br> | ||
| + | |||
| + | <b>2.CRY2-CIB1</b><br><br> | ||
| + | |||
| + | Cryptochromes (CRY) are photolyase-like blue-light receptors that mediate light responses in plants and animals. Arabidopsis CIB1 (cryptochrome-interacting basic-helix-loop-helix) protein interacts and only interacts with CRY2 (cryptochrome 2) under blue light condition. This has been varified in both yeast and Arabidopsis cells. Recently, a variety of microbial and plant-derived light-sensitive proteins have been engineered as optogenetic actuators, enabling high-precision spatiotemporal control of many cellular functions. The CRY2-CIB1 system has also been successfully applied to enable light-controlled gene expression in mammalian cells <i>in vivo</i>. <br><br> | ||
| + | |||
| + | <b>3.Our project</b><br><br> | ||
| + | |||
| + | Yeast-two-hybrid, the original method to discover the interaction between CRY2-CIB1, is now utilized in our project to enable temporal control of the target gene expression. Fusion proteins on one engineered plasmid, CRY2-GAL4 BD and CIB1-GAL4 AD are constitutively expressed. GAL4 BD is the binding domain of a transcriptional factor called GAL4. This domain can bind to UAS in the promoter. GAL4 AD is the activation domain of Gal4, it can recruit other transcription factors to intiate gene transcription. Under blue light condition, the interaction between CRY2-CIB1 takes place, so that GAL4 AD and GAL4 BD reunite as a functional GAL4 transcription factor to induce the expression of the CASP3 gene down stream to UAS promoter. CASP3 expression will then induce apoptosis [fig.7]<br><br> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2014/0/07/HNUDiscription7.jpg" alt="Discription7"/> <br> | ||
| + | Fig.7 Iron sensitive aborbing system <br> <br> | ||
| + | |||
| + | <b>Reference</b><br> | ||
| + | <ol> | ||
| + | <li>Spectral and Kinetic Properties of the Fet3 Protein from Saccharomyces cerevisiae, a Multinuclear Copper Ferroxidase Enzyme*. Richard F. Hassett‡, Daniel S. Yuan§¶, and Daniel J. Kosman‡i. THE JOURNAL OF BIOLOGICAL CHEMISTRY. Vol. 273, No. 36, Issue of September 4, pp. 23274–23282, 1998</li> | ||
| + | <li>The Fet3 Protein: A Multicopper Ferroxidase Essential to Iron Metabolism in Yeast. Daniel J. Kosman. Handbook of Copper Pharmacology and Toxicology. March 1, 2002</li> | ||
| + | <li>The copper-iron connection in biology: Structure of the metallo-oxidase Fet3p. Alexander B. Taylor*, Christopher S. Stoj†, Lynn Ziegler†, Daniel J. Kosman†, and P. John Hart*‡. PNAS October 25, 2005 vol. 102 no. 43 15459–15464.</li> | ||
| + | <li>A Permease-Oxidase Complex Involved in High-Affinity Iron Uptake in Yeast. </li> | ||
| + | <li>Robert Stearman, Daniel S. Yuan, Yuko Yamaguchi-Iwai, Richard D. Klausner andAndrew Dancis. Science, New Series, Vol. 271, No. 5255 (Mar. 15, 1996) </li> | ||
| + | <li>IRON METABOLISM IN AN IN VITRO MODEL OF CARDIAC ISCHEMIA: HYPOXIC INJURY AND PROTECTIVE STRATEGIES. Virginia Cozzolino. UNIVERSITA’ DEGLI STUDI DI NAPOLI</li> | ||
| + | <li>Post-transcriptional Regulation of the Yeast High Affinity Iron Transport System. M. Rosa Felice, Ivana De Domenico, Liangtao, Li, Diane McVey Ward, Beatrix Bartok, Giovanni Musci and Jerry Kaplan. J. Biol. Chem. 2005, 280:22181-22190.</li> | ||
| + | <li>Post-transcriptional regulation of gene expression in response to iron deficiency: co-ordinated metabolic reprogramming by yeast mRNA-binding proteins. Sandra V. Vergara and Dennis J. Thiele1. Biochemical Society Transactions (2008) Volume 36, part 5.</li> | ||
| + | <li>Translational repression by the human iron-regulatory factor (IRF) in Saccharomyces cerevisiae. Carla C.Oliveira, Britta Goossen1, Nilson l.T.Zanchin, John E.G.McCarthy*, Matthias W.Hentze1 and Renata Stripecke1. Nucleic Acids Research, 1993, Vol. 21, No. 23</li> | ||
| + | <li>Emerging roles of caspase-3 in apoptosis. Alan G. Porter*,1 and Reiner U. JaÈ nicke1. Cell Death and Differentiation (1999) 6, 99 ± 104. </li> | ||
| + | <li>Caspase-dependent apoptosis in yeast. Cristina Mazzoni, Claudio Falcone. Biochimica et Biophysica Acta 1783 (2008) 1320–1327.</li> | ||
| + | <li>Apoptosis in Yeasts. Martin Weinberger, Lakshmi Ramachandran and William C. Burhans. IUBMB Life, 55(8): 467–472, August 2003.</li> | ||
| + | <li>A Caspase-Related Protease Regulates Apoptosis in Yeast. Frank Madeo,1,7 Eva Herker,1 Corinna Maldener,1</li> | ||
| + | <li>Silke Wissing,1 Stephan La¨ chelt,1 Mark Herlan,2, Markus Fehr,3 Kirsten Lauber,4 Stephan J. Sigrist,5, Sebastian Wesselborg,4 and Kai-Uwe Fro¨ hlich6. Molecular Cell, Vol. 9, 911–917, April, 2002</li> | ||
| + | <li>Photoexcited CRY2 Interacts with CIB1 to Regulate Transcription and Floral Initiation in Arabidopsis. Hongtao Liu, Xuhong Yu, Kunwu Li, John Klejnot, Hongyun Yang, Dominique Lisiero, Chentao Lin*. SCIENCE VOL 322 5 DECEMBER 2008.</li> | ||
| + | <li>Optical control of mammalian endogenous</li> | ||
| + | <li>transcription and epigenetic states. Silvana Konermann1,2*, Mark D. Brigham1,2,3*, Alexandro Trevino1,2, Patrick D. Hsu1,2,4, Matthias Heidenreich1,2, Le Cong1,2,5, Randall J. Platt1,2, David A. Scott1,2, George M. Church1,6 & Feng Zhang1,2. doi:10.1038/nature12466</li> | ||
| + | </ol> | ||
| + | </br> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
<div id="cfoot"></div> | <div id="cfoot"></div> | ||
</div> | </div> | ||
<div id="cont_3"> | <div id="cont_3"> | ||
| - | <div id="chead"></div> | + | <div id="chead"> |
| - | <div id="cmiddle">3</div> | + | <div align="left"class="STYLE3" style="margin-left:50px;"><br/><br/><br/>Experiment</div> |
| + | </div> | ||
| + | <div id="cmiddle"> | ||
| + | <div align="left" class="STYLE2" style="margin-left:50px;margin-right:50px;"> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/e/ea/June.png" width="98" height="45" /> <br/> | ||
| + | Preliminary construction of the Cry2-CIB1 system <br/><br/> | ||
| + | |||
| + | <b>I.Cloning of the Cry2 & CIB1 gene</b> <br/><br/> | ||
| + | |||
| + | According to the sequences from NCBI, we designed two pairs of primers respectively, and cloned them based on the cDNA library of <i>Arabidopsis</i>. To fulfill the requirement of following gateway operation, the attB1/2 are added to the primers.<br/><br/> | ||
| + | |||
| + | Primer for Cry2: <br/> | ||
| + | Fwd 5’→3’ <span class="STYLE4">GTACAAAAAAGCAG</span>ATGAATGGAGCTATAGGA <br/> | ||
| + | attB1 <br/> | ||
| + | Rev 5’→3’ <span class="STYLE4">GTACAAGAAAGCTGGGT</span>TCAAACTCCTAAATTGCC <br/> | ||
| + | attB2 <br/><br/> | ||
| + | |||
| + | Primer for CIB1: <br/> | ||
| + | Fwd 5’→3’ <span class="STYLE4">GTACAAAAAAGCAG</span>ATGAAGATGGACAAAAAGA <br/> | ||
| + | attB1 <br/> | ||
| + | Rev 5’→3’ <span class="STYLE4">GTACAAGAAAGCTGGGT</span>TCATTTGCAACCATTTTT <br/> | ||
| + | attB1 <br/><br/> | ||
| + | |||
| + | <b>II.Yeast-two-hybrid </b> <br/><br/> | ||
| + | |||
| + | What we chose is the commercial shuttle vectors, pDEST32 and pDEST22[fig.1], where the DBD and AD is actually Gal4 BD and Gal4 AD respectively, and the Gal4 AD shall specifically bind to the upstream activating sequence(UAS), locating in the promoters of several reporter genes in the yeast strain genome like lacZ, HIS3 as well URA3.<br/><br/> | ||
| + | |||
| + | Fig.2 The map of our two plasmids: pDEST22 and pDEST32<br/> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/0/09/Experimentation_fig2.jpg" /><br/><br/> | ||
| + | |||
| + | Since we have obtained CRY2 and CIB1, what we do next is to induce both of them into their vectors with the technology named Gateway, constructing two new plasmids, pDEST22-CIB1 and pDEST32-32 as below[fig.2]. Then we transform the plasmids above into the Saccharomyces cerevisiae cell, AH109, screening by Leu and Trp auxotroph. <br/><br/> | ||
| + | |||
| + | Fig.3 The map of our two plasmids: pDEST22-CIB1 and pDEST32-CRY2<br/> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/d/d9/Experimentation_fig3.jpg" /><br/><br/> | ||
| + | |||
| + | <b>III. Optogenetic test</b> <br/><br/> | ||
| + | |||
| + | To test the effect of light switchable gene expression system, blue-white selection is induced when the blue light in special incubator[fig.3] shall initiate the expression of LacZ, which combined with the IPTG and X-gal in solid medium, makes the colony into blue, contrarily, colony in dark remains white.[fig.4]<br/><br/> | ||
| + | |||
| + | Fig.3 Blue light incubator<br/> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/1/19/Experimentation_fig4.jpg" /><br/><br/> | ||
| + | |||
| + | Fig.4 The positive result of yeast X-gal coloration<br/> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/b/b6/Exfig4.jpg" /><br/><br/> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2014/7/73/July.png" width="98" height="45" /> <br/> | ||
| + | |||
| + | Cloning of the genes we need and ligated them into of T-vector <br/><br/> | ||
| + | |||
| + | <b>I.IRE-IRP</b> <br/><br/> | ||
| + | |||
| + | 1.IRP <br/><br/> | ||
| + | |||
| + | We decide to clone the IRP from human cDNA library. <br/><br/> | ||
| + | |||
| + | Primers for IRP[fig.5]: <br/> | ||
| + | Fwd 5’→3’ GCG<span class="STYLE4">AAGCTT</span>TCAGTAATCATGAGCAAC <br/> | ||
| + | HindIII <br/> | ||
| + | Rev 5’→3’ GCG<span class="STYLE4">GAGCTC</span>TTGAGCAGAGCGTAAGA <br/> | ||
| + | SacI <br/><br/> | ||
| + | |||
| + | Fig.5<br/> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/4/4b/Jiao1.jpg" /><br/><br/> | ||
| + | |||
| + | 2.IRE <br/><br/> | ||
| + | |||
| + | Since IRE is a quite short sequence, counting 40 bp at all. A pair of single-stranded DNA are synthesized as the primers, and annealing shall create double-stranded DNA. <br/><br/> | ||
| + | |||
| + | Pair of single-stranded DNAs <br/> | ||
| + | Fwd 5’→3’ GCG<span class="STYLE4">AAGCTT</span>GTTCTTGCTTCAACAGTGTTTGGACGGAAC<span class="STYLE4">ACTAGT</span>GCG <br/> | ||
| + | HindIII SpeI <br/> | ||
| + | Rev 5’→3’ CGC<span class="STYLE4">ACTAGT</span>GTTCCGTCCAAACACTGTTGAAGCAAGAAC<span class="STYLE4">AAGCTT</span>CGC <br/> | ||
| + | SpeI HindIII <br/><br/> | ||
| + | |||
| + | <b>II.FET3</b> <br/><br/> | ||
| + | |||
| + | 1.Prepare for the genome of <i>Saccharomyces cerevisiae</i> <br/><br/> | ||
| + | |||
| + | Due to the lack of existing yeast genome as the PCR template, we order a yeast genome extraction kit to prepare the source.[fig.2] <br/><br/> | ||
| + | |||
| + | Fig.2 <br/> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/a/a7/Jiao2.jpg" /><br/><br/> | ||
| + | |||
| + | 2.Cloning of FET3 <br/><br/> | ||
| + | |||
| + | Primers for FET3[fig.3] <br/> | ||
| + | Fwd 5’→3’ GCG<span class="STYLE4">AAGCTT</span>ATGACTAACGCTTTGCTCTCTATAG <br/> | ||
| + | HindIII <br/> | ||
| + | Rev 5’→3’ GCG<span class="STYLE4">GAGCTC</span>TGGAACCCTTGACCGA <br/> | ||
| + | SacI <br/><br/> | ||
| + | |||
| + | Fig.3 <br/> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/1/11/Jiao3.jpg" /><br/><br/> | ||
| + | |||
| + | <b>III.CASP3</b> <br/><br/> | ||
| + | |||
| + | Caspase-3 normally exists in the cytosolic fraction of cells as an inactive precursor that is activated proteolytically when cells are signaled to undergo apoptosis.[4] So in the project, we want to make the modified yeast commit suicide through overexpress caspase-3 protein to protect natural environment. <br/><br/> | ||
| + | |||
| + | The template for PCR is also the human cDNA library. <br/><br/> | ||
| + | |||
| + | Primers for CASP3[fig.4] <br/> | ||
| + | Fwd 5’→3’ GCG<span class="STYLE4">ACTAGT</span>GCTCTGGTTTTCGGTGGG <br/> | ||
| + | Spe1 <br/> | ||
| + | Rev 5’→3’ GCG<span class="STYLE4">GAGCTC</span>TGGAACCCTTGACCGA <br/> | ||
| + | SacI <br/><br/> | ||
| + | |||
| + | Fig.4 <br/> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/1/18/Jiao4.jpg" /><br/><br/> | ||
| + | |||
| + | <b>IV.TA cloning</b> <br/><br/> | ||
| + | |||
| + | To make sure the PCR-cloned sequences are expected, we ligate them into the pMD18-T vector, then sequence all together. On the other hand, objective sequence in circular plasmids makes it convenient and stable for storage for quite long time. <br/><br/> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2014/a/a0/August.png" width="98" height="45" /> <br/> | ||
| + | |||
| + | Construction of the two plasmids in iron sensitive absorbing system<br/><br/> | ||
| + | |||
| + | Since we comfronted trouble in the ligation of gene with shuttle vector by T4 ligase after restriction enzyme treatment, the schematics below is just our original plan.<br/><br/> | ||
| + | |||
| + | <b>I.pDEST22-IRP1</b> <br/><br/> | ||
| + | |||
| + | Concerning the limitation of restriction cloning site, We have to ligated IRP1 firstly with pDEST32, and finally tranfer it into the pDEST22 vector.<br/><br/> | ||
| + | |||
| + | 1.When pDEST32 are treated with HindIII SacI, it yields segment of 2.3K & 10K.<br/> | ||
| + | 2.When T-IRP1 are treated with HindIII SacI, it yields segment of 2.6K.<br/> | ||
| + | 3.We can ligate the 10K and 2.6K segments together, constructing pDEST32-IREBP1.<br/> | ||
| + | 4.Restriction enzymens XhoI and SacI digest pDEST22 and pDEST32-IREBP1 respectively. <br/> | ||
| + | 5.The pDEST22-IREBP1 comes from the ligation of 5.1K segment from pDEST22 yields and the 4.1K segment from pDEST32-IREBP1 yields.<br/><br/> | ||
| + | |||
| + | <b>II.pDEST32-IREBP1</b> <br/><br/> | ||
| + | |||
| + | 1.Restriction enzymens HindIII and SacI digest pDEST32 and T-IRE respectively. <br/> | ||
| + | 2.pDEST32-IRE is the ligation product of 10K segment from pDEST32 and IRE<br/> | ||
| + | 3.Restriction enzymens SpeI and SacI digest pDEST32-IRE and T-FET3 respectively. <br/> | ||
| + | 4.pDEST32-IRE-FET3 is the ligation product of 10K segment from pDEST32-IRE and 2K segment from T-FET3<br/> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
<div id="cfoot"></div> | <div id="cfoot"></div> | ||
| - | + | </div> | |
| + | |||
<div id="cont_4"> | <div id="cont_4"> | ||
| - | <div id="chead"></div> | + | <div id="chead"> |
| - | <div id="cmiddle">4</div> | + | <div align="left"class="STYLE3" style="margin-left:50px;"><br/><br/><br/>Result</div> |
| + | </div> | ||
| + | <div id="cmiddle"> | ||
| + | <div align="left" class="STYLE2" style="margin-left:50px;margin-right:50px;"> | ||
| + | <br><b>Gene cloning</b><br><br> | ||
| + | |||
| + | The genes we have cloned:<br> | ||
| + | 1.IRP and CASP3 are from existing cDNA library of human.<br> | ||
| + | 2.CRY2, CIB1 are from existing cDNA library of <i>Arabidopsis</i>.<br> | ||
| + | 3.FET3 is from the genome of <i>Saccharomyces cerevisiae</i> made by ourselves.<br> | ||
| + | 4.IRE is made by annealing of two synthetic sigle-strain DNA.<br><br> | ||
| + | |||
| + | <b>TA cloning</b><br><br> | ||
| + | |||
| + | For sequencing and the stability of conservation, IRP, CASP3, FET3 and IRE are all introduced into the pMD-18T vector.<br><br> | ||
| + | |||
| + | <b>Optogenetic yeast-two-hybrid</b><br><br> | ||
| + | |||
| + | CRY2 and CIB1 are introduced into the commercial shuttle vectors, pDEST32 and pDEST22, by the gateway operation. Both of the pDEST22-CIB1 and pDEST32-CRY2 are transformed into the <i>Saccharomyces cerevisiae</i> cell AH109. Auxotroph screening and X-gal coloration prove the viability for the blue light to mediate CRY2-CIB1 interaction. | ||
| + | </div> | ||
| + | |||
| + | |||
| + | </div> | ||
<div id="cfoot"></div> | <div id="cfoot"></div> | ||
</div> | </div> | ||
<div id="cont_5"> | <div id="cont_5"> | ||
| - | <div id="chead"></div> | + | <div id="chead"> |
| - | <div id="cmiddle"> | + | <div align="left"class="STYLE3" style="margin-left:50px;"><br/><br/><br/>Future work</div> |
| + | </div> | ||
| + | <div id="cmiddle"> | ||
| + | <div align="left" class="STYLE2" style="margin-left:50px;margin-right:50px;"> | ||
| + | <br><b>Step I. </b><br><br> | ||
| + | |||
| + | When all of the schematic work finished, we are firstly going to verify the hypothesis of the apoptosis effect when caspase-3 expressed in yeast. If not, as previous plan B, caspase-8 will be tried in our situation.<br><br> | ||
| + | |||
| + | <b>Step II. </b><br><br> | ||
| + | |||
| + | The kinetic parameter of the light mediation, iron sensing and iron absorption are with worth to detect.<br><br> | ||
| + | |||
| + | <b>Step III.</b><br><br> | ||
| + | |||
| + | Though we would like to see them in only one microorganism clone, according to the original design, iron sensitive absorbing system and optogenetic apoptosis system consists of two engineered plasmids repectively. Four plasmids in total makes it impossible for integration. Further genetically engineering design and operation are necessary, mainly in system simplification. | ||
| + | </div> | ||
| + | |||
| + | </div> | ||
<div id="cfoot"></div> | <div id="cfoot"></div> | ||
</div> | </div> | ||
Latest revision as of 01:51, 18 October 2014
Background
Introduction
Biomining, is used to describe the novel approach in mining industry when microorganisms are applied to the extraction and recovery of precious and base metals from ores and concentrates[fig.1]. Technicially, it consists two branches, the bioleaching and biooxidation. The bioleaching strictly refers to the case when microorganisms are used to solubilize the metal. While biooxidation mainly focus on the pretreatment of target metals by bio-processsing minerals that occlude target metals, such as thiosulfate encompasses the gold.

Fig.1 Biomining is used to extract copper from copper ore.
Although the same biological process had been unknowingly used to extract metals at mine sites in, for example Spain, the UK and China, for several hundred years, the modern era of bioming began with the discovery of the bactreium, Thiobacillus ferrooxidans(now Acidithiobacillus ferrooxidans) in the mid-1940s. In 1958 Kennecott Mining Company patented the use of Thiobacillus ferrooxidans for copper extraction from waste rock dumps at the Bingham Canyon mine in Utah, USA. A recent survey illustrates that currently bioming is commercially praticed in the production of 15% of copper, 5% of gold.
Why biomine?
Bio-extractive techniques have to compete with alternative approaches for extracting metals from ores and concentrates. Some, such as pyrometallurgical technologies[fig.2] (ore roasting/smelting) have been refined over millennia and often represent major investments by mining companies, while others, such as pressure leaching, are more recent non-biological innovations. Main microorganisms involved in mineral oxidation processes are autotrophs, and the processes operate usually at atmospheric pressure and at relatively low temperatures (20–80 °C). Biomining is generally perceived as a much more environmentally benign (‘green’) approach, involving much lower temperatures (and hence energy costs) and smaller carbon footprints, which contrasts with current biomining operations, relied on the blasting and grinding of ore bodies, emitting large amounts of CO2, consuming 5% of total global energy production. Bio- processing also has niche advantages in two circumstances: first, when the ore or concentrate contains significant quantities of arsenic; second, for processing lowgrade and complex (polymetallic) ores.

Fig2. Pyrometallurgy
Configurations and microbiology for biomining
Engineering options for biomining have evolved from relatively inexpensive, partly controlled, irrigated dump or heap reactors to sophisticated, highly controlled and expensive stirred-tank reactors. Another distinguished way of bioming is the in situ mining, used extensively in Canada in the 1970s to recover uranium from worked out deep mines, which may well be the next major development in the mining sector. Mineral heaps and stirred tanks provide very different environments and challenges for mineral-leaching microorganisms, and different ‘optimal’ populations might be expected to emerge with similar target minerals depending on the reactor used.
The extreme physico-chemical nature of bioleach liquors — low pH, elevated concentrations of (toxic) metals, metalloids and other solutes, and highly positive redox potential (EH values may exceed +900 mV) — means that they are highly toxic to the vast majority of life forms, including microorganisms. Microorganisms oxidize both sulfur and iron of sulfide minerals, such as pyrite. It is now well established that bioleaching and biooxidation in all biomining operations is mediated by consortia of acidophilic prokaryotes. These have been categorized as: firstly, ferric iron-generating autotrophs which produce the mineral oxidant; secondly, sulfuric acid-generating autotrophs, which maintain the low pH environment required; and finally, heterotrophic and mixotropic prokaryotes, which degrade organic compounds leaked from autotrophic iron-oxidizers and sulfur-oxidizers, there avoiding potential toxicity issues.
Engineered microbial consortia
The field of synthetic biology has developed a wide range of highly engineered clonal populations of bacteria to perform complex tasks.The construction and analysis of synthetic gene circuits has not only provided us with new tools for genetic engineering but has given deeper insight into naturally occurring gene circuits, their evolution, architectures, and properties as well. The industrial practice of biomining and bioremediation of heavy metal contaminations could potentially benefit from synthetic consortia as natural consortia have been shown to play crucial roles in these processes.
To our best knowledge, hybrid consortia consisting of genetically engineered and naturally occurring bioleaching bacteria have not been reported so far. In fact only two knockouts and two expression mutants have been reported in the scientific literature. One rus overexpressing A. ferrooxidans strain and another expressing the mer determinant for a mercury resistant A. caldus strain. Once more suitable transformation protocols have been developed, it maybe feasible to modulate quorum-sensing signals with engineered microbes by either attenuating or amplifying natural signals or sending artificial signals to promote biofilm formation or mobilization respectively as recently demonstrated with engineered E.coli cells.
Fungi in acid mine drainage
Roles of fungi, the natural residents of acid mine drainage(AMD) and its attenuator are not emphasized adequately in the mine water research. Though AMD appears to be a certain special enviroment with relatively high metal concentration as well low pH, several species of fungi are also isolated from the AMD carrying streams, even AMD. Generally, fungi occur over a wide pH range (pH 1.0–11.0) and have been detected in acid habitats like volcanic springs, acid mine drainage or acid industrial wastewaters. Many of them are primarily acid-tolerant, but truly acidophilic species have also been detected[talbe.1]. While extracellular precipitation, complexation and crystallization, metal transformation, biosorption and sequestration are seen for fungi, endow fungi the ability to survive.

Under this condition, fungi may play the important role as primary degraders of complex organic matter, due to the absence of invertebrates that actively shred the leaves (shredders) at pH values below 3.5. At the same time, the fungi will contribute to oxygen consumption, thereby limiting oxidative stress for the anaerobic bactiria. Moreover, fungi can be directly involved in the reduction of ferric iron or sulphur. Comparatively, more reports are available concerning the absorption of heavy metals by fungi, in comparison to bacteria or algae in freshwater ecosystem. Fungi can absorb metals in their cell wall or adsorb in extracellular polysaccharide slime. This capacity enables them to grow in the presence of high amounts of heavy metals. Fungal activity in acid mine drainage is represented in Fig.3

Fig.3 Schematic diagram of fungal influence in acid mine drainage remediation
Conclusion
Biomining, is used to describe the novel approach in mining industry when microorganisms are applied to the extraction and recovery of precious and base metals from ores and concentrates. Compared with the conventional approachs, such as pyrometallurgical technologies and pressure leaching in mining industry, bioming has its unique advantages in energy conservation and emission reduction, both of which are definitely with rising importance in the modern time. Microorganisms are usually settled in the non-sterile condition, and participate the bio-processing in consortia. In most of the time, they play the role of iron-oxidizer, sulfur-oxdizer and mixotrophic or heterotrophic acidophiles repectively or together. In the “bottom up” approach to optimize the consortia, the logic of synthetic biology is a worthy try to get the optimum colonies. Fungi is a natural resident in the acid mine drainage and involved in the reduction of ferric iron and sulphur, matching the main role in bioming consortia. Among these fungi is Saccharomyces cerevisiae, one of the best studied species and a very popular model organism for synthetic biology. The goal of our project is to take advantage of synthetic biology approach to synthesize a optimized biomining Saccharomyces cerevisiae strain.
Reference
Biomining, is used to describe the novel approach in mining industry when microorganisms are applied to the extraction and recovery of precious and base metals from ores and concentrates[fig.1]. Technicially, it consists two branches, the bioleaching and biooxidation. The bioleaching strictly refers to the case when microorganisms are used to solubilize the metal. While biooxidation mainly focus on the pretreatment of target metals by bio-processsing minerals that occlude target metals, such as thiosulfate encompasses the gold.

Fig.1 Biomining is used to extract copper from copper ore.
Although the same biological process had been unknowingly used to extract metals at mine sites in, for example Spain, the UK and China, for several hundred years, the modern era of bioming began with the discovery of the bactreium, Thiobacillus ferrooxidans(now Acidithiobacillus ferrooxidans) in the mid-1940s. In 1958 Kennecott Mining Company patented the use of Thiobacillus ferrooxidans for copper extraction from waste rock dumps at the Bingham Canyon mine in Utah, USA. A recent survey illustrates that currently bioming is commercially praticed in the production of 15% of copper, 5% of gold.
Why biomine?
Bio-extractive techniques have to compete with alternative approaches for extracting metals from ores and concentrates. Some, such as pyrometallurgical technologies[fig.2] (ore roasting/smelting) have been refined over millennia and often represent major investments by mining companies, while others, such as pressure leaching, are more recent non-biological innovations. Main microorganisms involved in mineral oxidation processes are autotrophs, and the processes operate usually at atmospheric pressure and at relatively low temperatures (20–80 °C). Biomining is generally perceived as a much more environmentally benign (‘green’) approach, involving much lower temperatures (and hence energy costs) and smaller carbon footprints, which contrasts with current biomining operations, relied on the blasting and grinding of ore bodies, emitting large amounts of CO2, consuming 5% of total global energy production. Bio- processing also has niche advantages in two circumstances: first, when the ore or concentrate contains significant quantities of arsenic; second, for processing lowgrade and complex (polymetallic) ores.

Fig2. Pyrometallurgy
Configurations and microbiology for biomining
Engineering options for biomining have evolved from relatively inexpensive, partly controlled, irrigated dump or heap reactors to sophisticated, highly controlled and expensive stirred-tank reactors. Another distinguished way of bioming is the in situ mining, used extensively in Canada in the 1970s to recover uranium from worked out deep mines, which may well be the next major development in the mining sector. Mineral heaps and stirred tanks provide very different environments and challenges for mineral-leaching microorganisms, and different ‘optimal’ populations might be expected to emerge with similar target minerals depending on the reactor used.
The extreme physico-chemical nature of bioleach liquors — low pH, elevated concentrations of (toxic) metals, metalloids and other solutes, and highly positive redox potential (EH values may exceed +900 mV) — means that they are highly toxic to the vast majority of life forms, including microorganisms. Microorganisms oxidize both sulfur and iron of sulfide minerals, such as pyrite. It is now well established that bioleaching and biooxidation in all biomining operations is mediated by consortia of acidophilic prokaryotes. These have been categorized as: firstly, ferric iron-generating autotrophs which produce the mineral oxidant; secondly, sulfuric acid-generating autotrophs, which maintain the low pH environment required; and finally, heterotrophic and mixotropic prokaryotes, which degrade organic compounds leaked from autotrophic iron-oxidizers and sulfur-oxidizers, there avoiding potential toxicity issues.
Engineered microbial consortia
The field of synthetic biology has developed a wide range of highly engineered clonal populations of bacteria to perform complex tasks.The construction and analysis of synthetic gene circuits has not only provided us with new tools for genetic engineering but has given deeper insight into naturally occurring gene circuits, their evolution, architectures, and properties as well. The industrial practice of biomining and bioremediation of heavy metal contaminations could potentially benefit from synthetic consortia as natural consortia have been shown to play crucial roles in these processes.
To our best knowledge, hybrid consortia consisting of genetically engineered and naturally occurring bioleaching bacteria have not been reported so far. In fact only two knockouts and two expression mutants have been reported in the scientific literature. One rus overexpressing A. ferrooxidans strain and another expressing the mer determinant for a mercury resistant A. caldus strain. Once more suitable transformation protocols have been developed, it maybe feasible to modulate quorum-sensing signals with engineered microbes by either attenuating or amplifying natural signals or sending artificial signals to promote biofilm formation or mobilization respectively as recently demonstrated with engineered E.coli cells.
Fungi in acid mine drainage
Roles of fungi, the natural residents of acid mine drainage(AMD) and its attenuator are not emphasized adequately in the mine water research. Though AMD appears to be a certain special enviroment with relatively high metal concentration as well low pH, several species of fungi are also isolated from the AMD carrying streams, even AMD. Generally, fungi occur over a wide pH range (pH 1.0–11.0) and have been detected in acid habitats like volcanic springs, acid mine drainage or acid industrial wastewaters. Many of them are primarily acid-tolerant, but truly acidophilic species have also been detected[talbe.1]. While extracellular precipitation, complexation and crystallization, metal transformation, biosorption and sequestration are seen for fungi, endow fungi the ability to survive.

Under this condition, fungi may play the important role as primary degraders of complex organic matter, due to the absence of invertebrates that actively shred the leaves (shredders) at pH values below 3.5. At the same time, the fungi will contribute to oxygen consumption, thereby limiting oxidative stress for the anaerobic bactiria. Moreover, fungi can be directly involved in the reduction of ferric iron or sulphur. Comparatively, more reports are available concerning the absorption of heavy metals by fungi, in comparison to bacteria or algae in freshwater ecosystem. Fungi can absorb metals in their cell wall or adsorb in extracellular polysaccharide slime. This capacity enables them to grow in the presence of high amounts of heavy metals. Fungal activity in acid mine drainage is represented in Fig.3

Fig.3 Schematic diagram of fungal influence in acid mine drainage remediation
Conclusion
Biomining, is used to describe the novel approach in mining industry when microorganisms are applied to the extraction and recovery of precious and base metals from ores and concentrates. Compared with the conventional approachs, such as pyrometallurgical technologies and pressure leaching in mining industry, bioming has its unique advantages in energy conservation and emission reduction, both of which are definitely with rising importance in the modern time. Microorganisms are usually settled in the non-sterile condition, and participate the bio-processing in consortia. In most of the time, they play the role of iron-oxidizer, sulfur-oxdizer and mixotrophic or heterotrophic acidophiles repectively or together. In the “bottom up” approach to optimize the consortia, the logic of synthetic biology is a worthy try to get the optimum colonies. Fungi is a natural resident in the acid mine drainage and involved in the reduction of ferric iron and sulphur, matching the main role in bioming consortia. Among these fungi is Saccharomyces cerevisiae, one of the best studied species and a very popular model organism for synthetic biology. The goal of our project is to take advantage of synthetic biology approach to synthesize a optimized biomining Saccharomyces cerevisiae strain.
Reference
- Biomining — biotechnologies for extracting and recovering metals from ores and waste materials. D Barrie Johnson. Current Opinion in Biotechnology 2014, 30:24–31
- The microbiology of biomining: development and optimization of mineral-oxidizing microbial consortia. Douglas E. Rawlings1 and D. Barrie Johnson2.Microbiology (2007), 153, 315–324
- How will biomining be applied in future? C. L. BRIERLEY. Trans. Nonferrous Met. Soc. China 18(2008) 1302-1310
- Engineering microbial consortia to enhance biomining and bioremediation. Karl D. Brune andTravis S. Bayer* . Frontiers in Microbiology. 05 June 2012
- Occurrence and role of algae and fungi in acid mine drainage environment with special reference to metals and sulfate immobilization. Bidus Kanti Dasa, Arup Roya, Matthias Koschorreckb, Santi M. Mandalc, Katrin Wendt-Potthoffb, Jayanta Bhattacharyaa,*. water research 43 (2009)883–894
Description
I.Overview
Biomining refers to new technologies that microorganisms are applied to extract and recover metals from ores and concentrates. In mordant mining industry, the application of biomining is rapidly increased, since it has the advantage of energy conservation and emission reduction. In biomining, microorganisms are mainly used as iron-oxidizer or sulfur-oxidizer to solubilize either target metals or the unwanted minerals that occlude target metals. Microorganisms has also been applied to recover metal from the abandoned ore or slags, and remediate Acid Mine Drainage (AMD). Fungi are the natural residents of AMD. With the ability of ferric iron oxidation and absorption, they can also take part into the biomining process and the following metal reduction operation. Saccharomyces cerevisiae is one of the fungi species that has been identified in the AMD, yet its utility in biomining has largely been ignored. Here, we genetically engineer Saccharomyces cerevisiae to enhance its capacity of ferric iron oxidation and absorption. Meanwhile, we also introduce an inducible apoptosis system into the yeast so that unwanted or escape transgenic yeasts can be easily distroyed. Traditional method used to optimize microbial consortium for biomining is to inculate indigenous microflora into the reactor and screen for the better recombination. Compared with this so call “top down” method, our approach takes advantage of the powerful yeast genetic engineering to logically and systemically improve the desired traits.
We name our designed Saccharomyces cerevisiae strain ‘microbial miner’. This microbial miner consists of two parallel systems, iron sensitive absorbing system and optogenetic apoptosis system.
II.Iron sensitive aborbing system
1.FET3 protein
High affinity iron uptake in Saccharomyces cerevisiae requires FET3 protein. Localized on the external cell surface, FET3 protein is proposed to facilitate iron uptake by catalyzing the oxidation of Fe(II) to Fe(III) by O2, shown as below:
4Fe(II) + O2 + 4H+ → 4Fe(III) + 2H2O
The posttranslational insertion of four copper ions into FET3 protein is essential for its activity, thus linking copper and iron homeostasis together[fig.1]. In this model, Fe(III) is the substrate for the iron permease, encoded by FTR1 protein[fig2].

Fig.1 Ribbon diagram of FET3 protein. The T1, T2, and T3 copper atoms are shown in blue, green, and yellow, respectively. The three plastocyanin-like domains are shown as blue, green, and gold. Included in red are the carbohydrates resolvable in the electron density map.

Fig.2 FET3 and FTR1 proteins assemble into a complex in a cellular compartment early in the secretory pathway, most likely in the ER. The complex progresses to a post-Golgi compartment, where the CCC2 protein mediates copper delivery to FET3 protein. Finally, the copper-loaded F ET3 protein, presumably still complexed to FTR1 protein, is delivered to the plasma membrane and becomes competent for iron transport.
2.IRE-IRP1
Saccharomyces cerevisiae post-translationally regulates the expression of the plasma membrane high affinity iron transport system, consisting FET3 protein and FTR1 protein, in response to iron need. Incubation of cells in high iron leads to the internalization and degradation of both FET3 protein and FTR1 protein. Since we are looking for a way to accumulate iron within the cell, the cell should promote the absorption with the presence of iron. We thus cast our eyes to other mammalian iron regulation system.
In human cell, the cellular levels of ferritin and transferrin receptor (TfR) are also primarily regulated at translational level upon changes in iron availability, through interactions between iron regulatory proteins (IRP1 and IRP2) and iron responsive elements (IRE) in the 5’ UTR of H- and L-ferritin mRNA and the 3’ UTR of TfR mRNA. When intracellular iron levels rise, IRP1 can’t bind IRE, IRP2 is degraded and ferritin mRNA is efficiently translated. [fig.3] When iron level is high, IRP1 exists as a cytosolic aconitase. When iron is low, it assumes an open configuration associated with the loss of iron atoms in the iron-sulfur cluster, and can bind the IRE stem loop.[fig.4]

Fig.3 Decreased iron supply activates binding of IRPs to IRE resulting in translational inhibition of the mRNAs encoding ferritin and stabilization of the Tf R mRNA. During high iron conditions, IRPs lose their affinity for IREs, increasing translation of ferritin mRNAs and degradation of the Tf R mRNA.

Fig.4 The Fe-S cluster in aconitase.
3.Our project
We plan to introduce IRE-IRP1 system into Saccharomyces cerevisiae to control the translation of FET3 mRNA, enhancing its iron aborption capability. In the circumstance of low iron concentration, the constitutively expressing IRP1 bonds to the stem loop of IRE preceding the FET3 mRNA, impeding the translation of FET3. Once iron reaches to a high level, the breakdown of IRE-IRP1 interaction shall activate the FET3 translation, which lead to more iron uptake, at last achieve the goal for iron accumulation.[fig.5]

Fig.5 Iron sensitive aborbing system
III.Optogenetic apoptosis system
1.Caspase-3
Apoptosis or programmed cell death (PCD) is a highly coordinated cellular suicide program. Caspases, cysteine-dependent aspartate-directed proteases are a family of cysteine proteases that play essential roles in apoptosis[fig.6]. In previous studies in yeast, only the over-expression of caspase 8 led to cell death while the over-expression of caspase 3 was not lethal and only retarded yeast growth. In our design, the apoptosis related protein expresses when exposed to the blue light. Totally dark is harsh to fulfill in the industrial practicing, and urge us put the possibility of expression leakage into consideration, when the toxity of caspase may interfere the iron uptaking. Hence, we choose caspase-3, instead of caspase-8, as the apoptosis trigger.

Fig.6 Extrinsic and intrinsic pathways to caspase-3 activation.
Although yeasts lack some elements of the complex apoptotic machinery of metazoan cells, recent studies show that many features of apoptosis, including a caspase-like activity, can be induced in these organisms by DNA damage and other apoptotic triggers. Here we plan to introduce CASP3 gene from human, and attempt to verify our hypothesis of the apoptosis effect in yeast by Caspase-3.
2.CRY2-CIB1
Cryptochromes (CRY) are photolyase-like blue-light receptors that mediate light responses in plants and animals. Arabidopsis CIB1 (cryptochrome-interacting basic-helix-loop-helix) protein interacts and only interacts with CRY2 (cryptochrome 2) under blue light condition. This has been varified in both yeast and Arabidopsis cells. Recently, a variety of microbial and plant-derived light-sensitive proteins have been engineered as optogenetic actuators, enabling high-precision spatiotemporal control of many cellular functions. The CRY2-CIB1 system has also been successfully applied to enable light-controlled gene expression in mammalian cells in vivo.
3.Our project
Yeast-two-hybrid, the original method to discover the interaction between CRY2-CIB1, is now utilized in our project to enable temporal control of the target gene expression. Fusion proteins on one engineered plasmid, CRY2-GAL4 BD and CIB1-GAL4 AD are constitutively expressed. GAL4 BD is the binding domain of a transcriptional factor called GAL4. This domain can bind to UAS in the promoter. GAL4 AD is the activation domain of Gal4, it can recruit other transcription factors to intiate gene transcription. Under blue light condition, the interaction between CRY2-CIB1 takes place, so that GAL4 AD and GAL4 BD reunite as a functional GAL4 transcription factor to induce the expression of the CASP3 gene down stream to UAS promoter. CASP3 expression will then induce apoptosis [fig.7]

Fig.7 Iron sensitive aborbing system
Reference
Biomining refers to new technologies that microorganisms are applied to extract and recover metals from ores and concentrates. In mordant mining industry, the application of biomining is rapidly increased, since it has the advantage of energy conservation and emission reduction. In biomining, microorganisms are mainly used as iron-oxidizer or sulfur-oxidizer to solubilize either target metals or the unwanted minerals that occlude target metals. Microorganisms has also been applied to recover metal from the abandoned ore or slags, and remediate Acid Mine Drainage (AMD). Fungi are the natural residents of AMD. With the ability of ferric iron oxidation and absorption, they can also take part into the biomining process and the following metal reduction operation. Saccharomyces cerevisiae is one of the fungi species that has been identified in the AMD, yet its utility in biomining has largely been ignored. Here, we genetically engineer Saccharomyces cerevisiae to enhance its capacity of ferric iron oxidation and absorption. Meanwhile, we also introduce an inducible apoptosis system into the yeast so that unwanted or escape transgenic yeasts can be easily distroyed. Traditional method used to optimize microbial consortium for biomining is to inculate indigenous microflora into the reactor and screen for the better recombination. Compared with this so call “top down” method, our approach takes advantage of the powerful yeast genetic engineering to logically and systemically improve the desired traits.
We name our designed Saccharomyces cerevisiae strain ‘microbial miner’. This microbial miner consists of two parallel systems, iron sensitive absorbing system and optogenetic apoptosis system.
II.Iron sensitive aborbing system
1.FET3 protein
High affinity iron uptake in Saccharomyces cerevisiae requires FET3 protein. Localized on the external cell surface, FET3 protein is proposed to facilitate iron uptake by catalyzing the oxidation of Fe(II) to Fe(III) by O2, shown as below:
4Fe(II) + O2 + 4H+ → 4Fe(III) + 2H2O
The posttranslational insertion of four copper ions into FET3 protein is essential for its activity, thus linking copper and iron homeostasis together[fig.1]. In this model, Fe(III) is the substrate for the iron permease, encoded by FTR1 protein[fig2].

Fig.1 Ribbon diagram of FET3 protein. The T1, T2, and T3 copper atoms are shown in blue, green, and yellow, respectively. The three plastocyanin-like domains are shown as blue, green, and gold. Included in red are the carbohydrates resolvable in the electron density map.

Fig.2 FET3 and FTR1 proteins assemble into a complex in a cellular compartment early in the secretory pathway, most likely in the ER. The complex progresses to a post-Golgi compartment, where the CCC2 protein mediates copper delivery to FET3 protein. Finally, the copper-loaded F ET3 protein, presumably still complexed to FTR1 protein, is delivered to the plasma membrane and becomes competent for iron transport.
2.IRE-IRP1
Saccharomyces cerevisiae post-translationally regulates the expression of the plasma membrane high affinity iron transport system, consisting FET3 protein and FTR1 protein, in response to iron need. Incubation of cells in high iron leads to the internalization and degradation of both FET3 protein and FTR1 protein. Since we are looking for a way to accumulate iron within the cell, the cell should promote the absorption with the presence of iron. We thus cast our eyes to other mammalian iron regulation system.
In human cell, the cellular levels of ferritin and transferrin receptor (TfR) are also primarily regulated at translational level upon changes in iron availability, through interactions between iron regulatory proteins (IRP1 and IRP2) and iron responsive elements (IRE) in the 5’ UTR of H- and L-ferritin mRNA and the 3’ UTR of TfR mRNA. When intracellular iron levels rise, IRP1 can’t bind IRE, IRP2 is degraded and ferritin mRNA is efficiently translated. [fig.3] When iron level is high, IRP1 exists as a cytosolic aconitase. When iron is low, it assumes an open configuration associated with the loss of iron atoms in the iron-sulfur cluster, and can bind the IRE stem loop.[fig.4]

Fig.3 Decreased iron supply activates binding of IRPs to IRE resulting in translational inhibition of the mRNAs encoding ferritin and stabilization of the Tf R mRNA. During high iron conditions, IRPs lose their affinity for IREs, increasing translation of ferritin mRNAs and degradation of the Tf R mRNA.

Fig.4 The Fe-S cluster in aconitase.
3.Our project
We plan to introduce IRE-IRP1 system into Saccharomyces cerevisiae to control the translation of FET3 mRNA, enhancing its iron aborption capability. In the circumstance of low iron concentration, the constitutively expressing IRP1 bonds to the stem loop of IRE preceding the FET3 mRNA, impeding the translation of FET3. Once iron reaches to a high level, the breakdown of IRE-IRP1 interaction shall activate the FET3 translation, which lead to more iron uptake, at last achieve the goal for iron accumulation.[fig.5]

Fig.5 Iron sensitive aborbing system
III.Optogenetic apoptosis system
1.Caspase-3
Apoptosis or programmed cell death (PCD) is a highly coordinated cellular suicide program. Caspases, cysteine-dependent aspartate-directed proteases are a family of cysteine proteases that play essential roles in apoptosis[fig.6]. In previous studies in yeast, only the over-expression of caspase 8 led to cell death while the over-expression of caspase 3 was not lethal and only retarded yeast growth. In our design, the apoptosis related protein expresses when exposed to the blue light. Totally dark is harsh to fulfill in the industrial practicing, and urge us put the possibility of expression leakage into consideration, when the toxity of caspase may interfere the iron uptaking. Hence, we choose caspase-3, instead of caspase-8, as the apoptosis trigger.

Fig.6 Extrinsic and intrinsic pathways to caspase-3 activation.
Although yeasts lack some elements of the complex apoptotic machinery of metazoan cells, recent studies show that many features of apoptosis, including a caspase-like activity, can be induced in these organisms by DNA damage and other apoptotic triggers. Here we plan to introduce CASP3 gene from human, and attempt to verify our hypothesis of the apoptosis effect in yeast by Caspase-3.
2.CRY2-CIB1
Cryptochromes (CRY) are photolyase-like blue-light receptors that mediate light responses in plants and animals. Arabidopsis CIB1 (cryptochrome-interacting basic-helix-loop-helix) protein interacts and only interacts with CRY2 (cryptochrome 2) under blue light condition. This has been varified in both yeast and Arabidopsis cells. Recently, a variety of microbial and plant-derived light-sensitive proteins have been engineered as optogenetic actuators, enabling high-precision spatiotemporal control of many cellular functions. The CRY2-CIB1 system has also been successfully applied to enable light-controlled gene expression in mammalian cells in vivo.
3.Our project
Yeast-two-hybrid, the original method to discover the interaction between CRY2-CIB1, is now utilized in our project to enable temporal control of the target gene expression. Fusion proteins on one engineered plasmid, CRY2-GAL4 BD and CIB1-GAL4 AD are constitutively expressed. GAL4 BD is the binding domain of a transcriptional factor called GAL4. This domain can bind to UAS in the promoter. GAL4 AD is the activation domain of Gal4, it can recruit other transcription factors to intiate gene transcription. Under blue light condition, the interaction between CRY2-CIB1 takes place, so that GAL4 AD and GAL4 BD reunite as a functional GAL4 transcription factor to induce the expression of the CASP3 gene down stream to UAS promoter. CASP3 expression will then induce apoptosis [fig.7]

Fig.7 Iron sensitive aborbing system
Reference
- Spectral and Kinetic Properties of the Fet3 Protein from Saccharomyces cerevisiae, a Multinuclear Copper Ferroxidase Enzyme*. Richard F. Hassett‡, Daniel S. Yuan§¶, and Daniel J. Kosman‡i. THE JOURNAL OF BIOLOGICAL CHEMISTRY. Vol. 273, No. 36, Issue of September 4, pp. 23274–23282, 1998
- The Fet3 Protein: A Multicopper Ferroxidase Essential to Iron Metabolism in Yeast. Daniel J. Kosman. Handbook of Copper Pharmacology and Toxicology. March 1, 2002
- The copper-iron connection in biology: Structure of the metallo-oxidase Fet3p. Alexander B. Taylor*, Christopher S. Stoj†, Lynn Ziegler†, Daniel J. Kosman†, and P. John Hart*‡. PNAS October 25, 2005 vol. 102 no. 43 15459–15464.
- A Permease-Oxidase Complex Involved in High-Affinity Iron Uptake in Yeast.
- Robert Stearman, Daniel S. Yuan, Yuko Yamaguchi-Iwai, Richard D. Klausner andAndrew Dancis. Science, New Series, Vol. 271, No. 5255 (Mar. 15, 1996)
- IRON METABOLISM IN AN IN VITRO MODEL OF CARDIAC ISCHEMIA: HYPOXIC INJURY AND PROTECTIVE STRATEGIES. Virginia Cozzolino. UNIVERSITA’ DEGLI STUDI DI NAPOLI
- Post-transcriptional Regulation of the Yeast High Affinity Iron Transport System. M. Rosa Felice, Ivana De Domenico, Liangtao, Li, Diane McVey Ward, Beatrix Bartok, Giovanni Musci and Jerry Kaplan. J. Biol. Chem. 2005, 280:22181-22190.
- Post-transcriptional regulation of gene expression in response to iron deficiency: co-ordinated metabolic reprogramming by yeast mRNA-binding proteins. Sandra V. Vergara and Dennis J. Thiele1. Biochemical Society Transactions (2008) Volume 36, part 5.
- Translational repression by the human iron-regulatory factor (IRF) in Saccharomyces cerevisiae. Carla C.Oliveira, Britta Goossen1, Nilson l.T.Zanchin, John E.G.McCarthy*, Matthias W.Hentze1 and Renata Stripecke1. Nucleic Acids Research, 1993, Vol. 21, No. 23
- Emerging roles of caspase-3 in apoptosis. Alan G. Porter*,1 and Reiner U. JaÈ nicke1. Cell Death and Differentiation (1999) 6, 99 ± 104.
- Caspase-dependent apoptosis in yeast. Cristina Mazzoni, Claudio Falcone. Biochimica et Biophysica Acta 1783 (2008) 1320–1327.
- Apoptosis in Yeasts. Martin Weinberger, Lakshmi Ramachandran and William C. Burhans. IUBMB Life, 55(8): 467–472, August 2003.
- A Caspase-Related Protease Regulates Apoptosis in Yeast. Frank Madeo,1,7 Eva Herker,1 Corinna Maldener,1
- Silke Wissing,1 Stephan La¨ chelt,1 Mark Herlan,2, Markus Fehr,3 Kirsten Lauber,4 Stephan J. Sigrist,5, Sebastian Wesselborg,4 and Kai-Uwe Fro¨ hlich6. Molecular Cell, Vol. 9, 911–917, April, 2002
- Photoexcited CRY2 Interacts with CIB1 to Regulate Transcription and Floral Initiation in Arabidopsis. Hongtao Liu, Xuhong Yu, Kunwu Li, John Klejnot, Hongyun Yang, Dominique Lisiero, Chentao Lin*. SCIENCE VOL 322 5 DECEMBER 2008.
- Optical control of mammalian endogenous
- transcription and epigenetic states. Silvana Konermann1,2*, Mark D. Brigham1,2,3*, Alexandro Trevino1,2, Patrick D. Hsu1,2,4, Matthias Heidenreich1,2, Le Cong1,2,5, Randall J. Platt1,2, David A. Scott1,2, George M. Church1,6 & Feng Zhang1,2. doi:10.1038/nature12466
Experiment
Preliminary construction of the Cry2-CIB1 system
I.Cloning of the Cry2 & CIB1 gene
According to the sequences from NCBI, we designed two pairs of primers respectively, and cloned them based on the cDNA library of Arabidopsis. To fulfill the requirement of following gateway operation, the attB1/2 are added to the primers.
Primer for Cry2:
Fwd 5’→3’ GTACAAAAAAGCAGATGAATGGAGCTATAGGA
attB1
Rev 5’→3’ GTACAAGAAAGCTGGGTTCAAACTCCTAAATTGCC
attB2
Primer for CIB1:
Fwd 5’→3’ GTACAAAAAAGCAGATGAAGATGGACAAAAAGA
attB1
Rev 5’→3’ GTACAAGAAAGCTGGGTTCATTTGCAACCATTTTT
attB1
II.Yeast-two-hybrid
What we chose is the commercial shuttle vectors, pDEST32 and pDEST22[fig.1], where the DBD and AD is actually Gal4 BD and Gal4 AD respectively, and the Gal4 AD shall specifically bind to the upstream activating sequence(UAS), locating in the promoters of several reporter genes in the yeast strain genome like lacZ, HIS3 as well URA3.
Fig.2 The map of our two plasmids: pDEST22 and pDEST32

Since we have obtained CRY2 and CIB1, what we do next is to induce both of them into their vectors with the technology named Gateway, constructing two new plasmids, pDEST22-CIB1 and pDEST32-32 as below[fig.2]. Then we transform the plasmids above into the Saccharomyces cerevisiae cell, AH109, screening by Leu and Trp auxotroph.
Fig.3 The map of our two plasmids: pDEST22-CIB1 and pDEST32-CRY2

III. Optogenetic test
To test the effect of light switchable gene expression system, blue-white selection is induced when the blue light in special incubator[fig.3] shall initiate the expression of LacZ, which combined with the IPTG and X-gal in solid medium, makes the colony into blue, contrarily, colony in dark remains white.[fig.4]
Fig.3 Blue light incubator

Fig.4 The positive result of yeast X-gal coloration

Cloning of the genes we need and ligated them into of T-vector
I.IRE-IRP
1.IRP
We decide to clone the IRP from human cDNA library.
Primers for IRP[fig.5]:
Fwd 5’→3’ GCGAAGCTTTCAGTAATCATGAGCAAC
HindIII
Rev 5’→3’ GCGGAGCTCTTGAGCAGAGCGTAAGA
SacI
Fig.5

2.IRE
Since IRE is a quite short sequence, counting 40 bp at all. A pair of single-stranded DNA are synthesized as the primers, and annealing shall create double-stranded DNA.
Pair of single-stranded DNAs
Fwd 5’→3’ GCGAAGCTTGTTCTTGCTTCAACAGTGTTTGGACGGAACACTAGTGCG
HindIII SpeI
Rev 5’→3’ CGCACTAGTGTTCCGTCCAAACACTGTTGAAGCAAGAACAAGCTTCGC
SpeI HindIII
II.FET3
1.Prepare for the genome of Saccharomyces cerevisiae
Due to the lack of existing yeast genome as the PCR template, we order a yeast genome extraction kit to prepare the source.[fig.2]
Fig.2
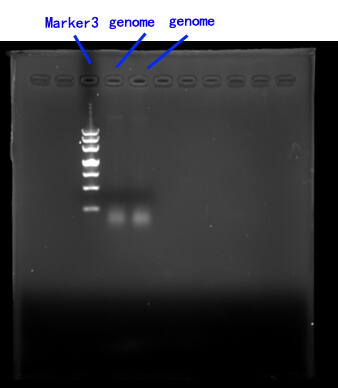
2.Cloning of FET3
Primers for FET3[fig.3]
Fwd 5’→3’ GCGAAGCTTATGACTAACGCTTTGCTCTCTATAG
HindIII
Rev 5’→3’ GCGGAGCTCTGGAACCCTTGACCGA
SacI
Fig.3
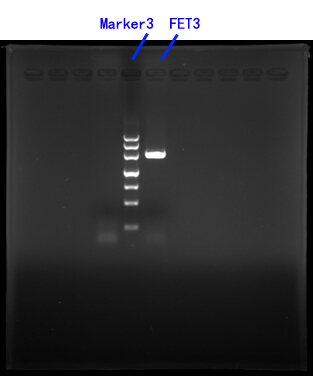
III.CASP3
Caspase-3 normally exists in the cytosolic fraction of cells as an inactive precursor that is activated proteolytically when cells are signaled to undergo apoptosis.[4] So in the project, we want to make the modified yeast commit suicide through overexpress caspase-3 protein to protect natural environment.
The template for PCR is also the human cDNA library.
Primers for CASP3[fig.4]
Fwd 5’→3’ GCGACTAGTGCTCTGGTTTTCGGTGGG
Spe1
Rev 5’→3’ GCGGAGCTCTGGAACCCTTGACCGA
SacI
Fig.4

IV.TA cloning
To make sure the PCR-cloned sequences are expected, we ligate them into the pMD18-T vector, then sequence all together. On the other hand, objective sequence in circular plasmids makes it convenient and stable for storage for quite long time.
Construction of the two plasmids in iron sensitive absorbing system
Since we comfronted trouble in the ligation of gene with shuttle vector by T4 ligase after restriction enzyme treatment, the schematics below is just our original plan.
I.pDEST22-IRP1
Concerning the limitation of restriction cloning site, We have to ligated IRP1 firstly with pDEST32, and finally tranfer it into the pDEST22 vector.
1.When pDEST32 are treated with HindIII SacI, it yields segment of 2.3K & 10K.
2.When T-IRP1 are treated with HindIII SacI, it yields segment of 2.6K.
3.We can ligate the 10K and 2.6K segments together, constructing pDEST32-IREBP1.
4.Restriction enzymens XhoI and SacI digest pDEST22 and pDEST32-IREBP1 respectively.
5.The pDEST22-IREBP1 comes from the ligation of 5.1K segment from pDEST22 yields and the 4.1K segment from pDEST32-IREBP1 yields.
II.pDEST32-IREBP1
1.Restriction enzymens HindIII and SacI digest pDEST32 and T-IRE respectively.
2.pDEST32-IRE is the ligation product of 10K segment from pDEST32 and IRE
3.Restriction enzymens SpeI and SacI digest pDEST32-IRE and T-FET3 respectively.
4.pDEST32-IRE-FET3 is the ligation product of 10K segment from pDEST32-IRE and 2K segment from T-FET3
Result
Gene cloning
The genes we have cloned:
1.IRP and CASP3 are from existing cDNA library of human.
2.CRY2, CIB1 are from existing cDNA library of Arabidopsis.
3.FET3 is from the genome of Saccharomyces cerevisiae made by ourselves.
4.IRE is made by annealing of two synthetic sigle-strain DNA.
TA cloning
For sequencing and the stability of conservation, IRP, CASP3, FET3 and IRE are all introduced into the pMD-18T vector.
Optogenetic yeast-two-hybrid
CRY2 and CIB1 are introduced into the commercial shuttle vectors, pDEST32 and pDEST22, by the gateway operation. Both of the pDEST22-CIB1 and pDEST32-CRY2 are transformed into the Saccharomyces cerevisiae cell AH109. Auxotroph screening and X-gal coloration prove the viability for the blue light to mediate CRY2-CIB1 interaction.
Future work
Step I.
When all of the schematic work finished, we are firstly going to verify the hypothesis of the apoptosis effect when caspase-3 expressed in yeast. If not, as previous plan B, caspase-8 will be tried in our situation.
Step II.
The kinetic parameter of the light mediation, iron sensing and iron absorption are with worth to detect.
Step III.
Though we would like to see them in only one microorganism clone, according to the original design, iron sensitive absorbing system and optogenetic apoptosis system consists of two engineered plasmids repectively. Four plasmids in total makes it impossible for integration. Further genetically engineering design and operation are necessary, mainly in system simplification.
 "
"







