Team:Brasil-SP/Project/ResponseModule
From 2014.igem.org
| (46 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | {{:Team:Brasil-SP/Templates/Header|headersrc=TheProjectBRASILSP.png}} | |
| - | < | + | <h1>Response Module</h1> |
| - | + | The reporter gene chosen for the output system was the GFP (REMINGTON, 2014). The main reason for that choice was the simplicity in measuring its fluorescenchttp with the fluorimeter and flow cytometry, but it can be replaced by any other reporter system. In the final development stage of the project we envision an output that do not need to be optically excited like the GFP does. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | </ | + | {{:Team:Brasil-SP/Templates/Image | image=Circuit_response.png | alignment=center | caption=. | size=500px}} |
| + | |||
| + | {{:Team:Brasil-SP/Templates/Image | image=Indicator.png | alignment=center | caption=. | size=500px}} | ||
| + | |||
| + | Note that the biossensor performs a negative detection. In other words, when the patient is healthy the bacteria will display a green fluorescence, and when in a unhealthy situation the bacteria will not glow. In a more realistic approach, we do not expect our system to have only two intensities (zero or maximum glow). What might happen is that we'll have a more continuous spectrum of intensities, which would make it harder to differentiate between a negative and positive diagnosis. To deal with this problem we are adding a intensity pattern ruler in our [https://2014.igem.org/Team:Brasil-SP/Project/Device microfluidic device]. Comparing the intensity displayed in the diagnosis chamber with the standardized ones the user will be able to analyse the result correctly | ||
| + | |||
| + | |||
| + | {{:Team:Brasil-SP/Templates/Image | image=GFP_cell.JPG | alignment=center | caption=High Intensity of GFP. | size=500px}} | ||
| + | |||
| + | {{:Team:Brasil-SP/Templates/Image | image=No_GFP.jpg | alignment=center | caption=Low/or no Intensity of GFP. | size=300px}} | ||
| + | |||
| + | <h2>Reference</h2> | ||
| + | #REMINGTON SJ. Green fluorescent protein: A perspective. <b>Protein Science</b>, 2011, 20(9):1509–1519. | ||
| + | |||
| + | |||
| + | {{:Team:Brasil-SP/Templates/Footer}} | ||
Latest revision as of 00:35, 18 October 2014
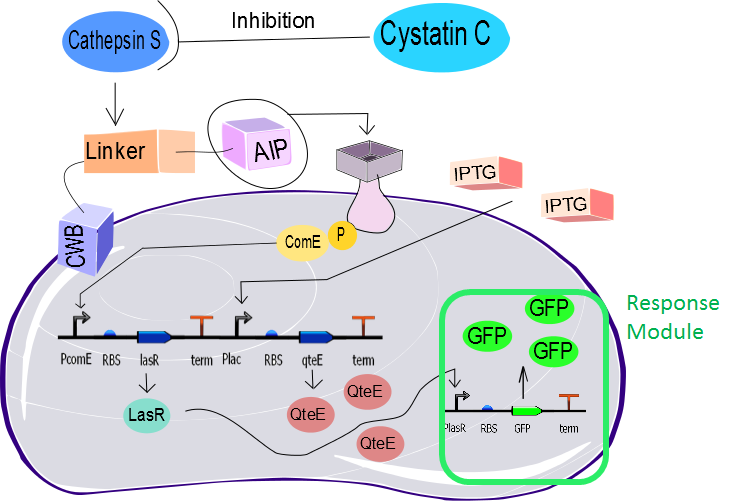
Response Module
The reporter gene chosen for the output system was the GFP (REMINGTON, 2014). The main reason for that choice was the simplicity in measuring its fluorescenchttp with the fluorimeter and flow cytometry, but it can be replaced by any other reporter system. In the final development stage of the project we envision an output that do not need to be optically excited like the GFP does.
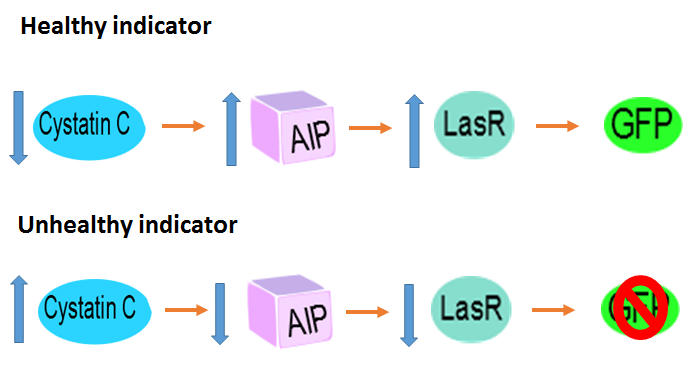
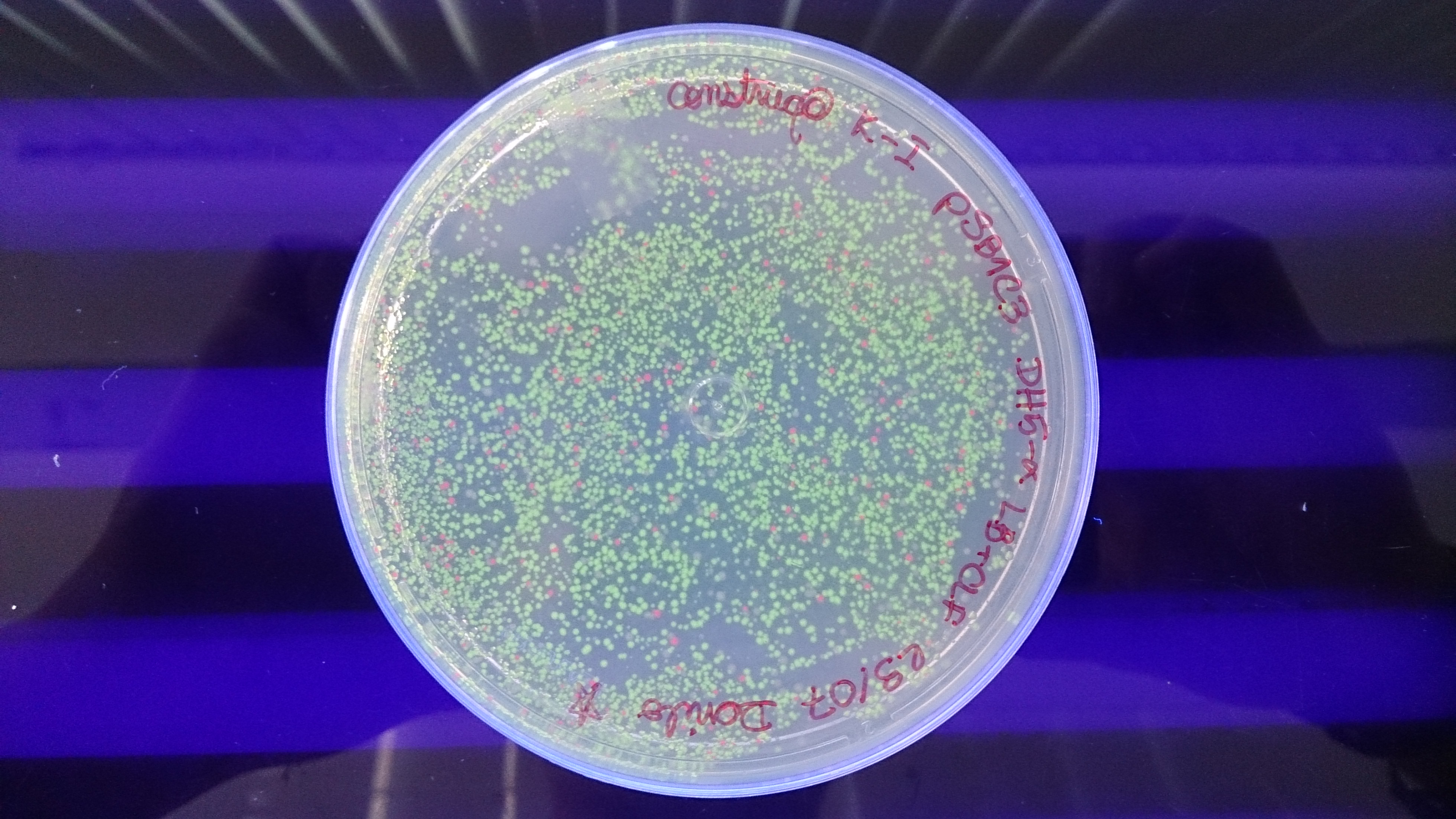
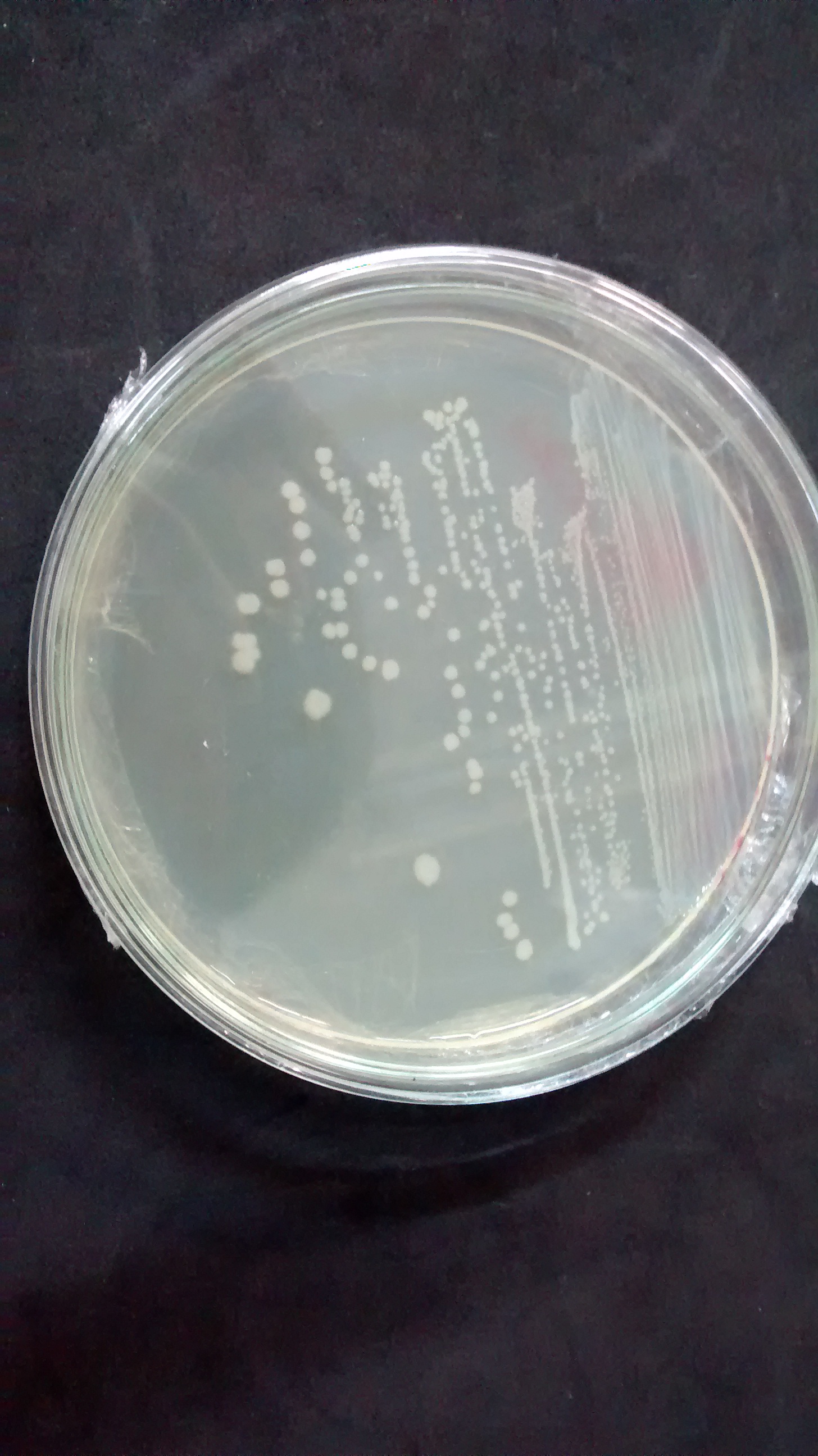
Note that the biossensor performs a negative detection. In other words, when the patient is healthy the bacteria will display a green fluorescence, and when in a unhealthy situation the bacteria will not glow. In a more realistic approach, we do not expect our system to have only two intensities (zero or maximum glow). What might happen is that we'll have a more continuous spectrum of intensities, which would make it harder to differentiate between a negative and positive diagnosis. To deal with this problem we are adding a intensity pattern ruler in our microfluidic device. Comparing the intensity displayed in the diagnosis chamber with the standardized ones the user will be able to analyse the result correctly
Reference
- REMINGTON SJ. Green fluorescent protein: A perspective. Protein Science, 2011, 20(9):1509–1519.
 "
"