Team:Heidelberg/pages/Xylanase
From 2014.igem.org
(→Design and cloning of constructs) |
(→Design and cloning of constructs) |
||
| Line 39: | Line 39: | ||
EcoRI site, the RBS [http://parts.igem.org/Part:BBa_B0034 B0034], a start codon (forward) and a SpeI site and a PstI site (reverse) were added by primer overhangs. The PCR product was cut with X and P and ligated into the expression vector [http://parts.igem.org/Part:BBa_K1362093 pSBX1K30]. In order to create the constructs to express linear xylanase with His6-Tag [http://parts.igem.org/Part:BBa_K1362023 BBa_K1362023] and linear xylanase with His6-tag and exteins, primers to linearize [http://parts.igem.org/Part:BBa_K1362020 BBa_K1362020] between the xylanase coding sequence and the stop codon and add BsaI sites at the ends were designed. Inserts coding for the sequence GSHHHHHHSG (His6-Tag) respectively HHHHHHRGKCWE (His6-tag + exteins) with overhangs corresponding to the overhangs caused by BsaI restriction of the vector PCR product were designed and ordered as DNA-oligomers. The inserts were annealed and inserted into the purified PCR product of [http://parts.igem.org/Part:BBa_K1362020 BBa_K1362020] by Golden Gate cloning. | EcoRI site, the RBS [http://parts.igem.org/Part:BBa_B0034 B0034], a start codon (forward) and a SpeI site and a PstI site (reverse) were added by primer overhangs. The PCR product was cut with X and P and ligated into the expression vector [http://parts.igem.org/Part:BBa_K1362093 pSBX1K30]. In order to create the constructs to express linear xylanase with His6-Tag [http://parts.igem.org/Part:BBa_K1362023 BBa_K1362023] and linear xylanase with His6-tag and exteins, primers to linearize [http://parts.igem.org/Part:BBa_K1362020 BBa_K1362020] between the xylanase coding sequence and the stop codon and add BsaI sites at the ends were designed. Inserts coding for the sequence GSHHHHHHSG (His6-Tag) respectively HHHHHHRGKCWE (His6-tag + exteins) with overhangs corresponding to the overhangs caused by BsaI restriction of the vector PCR product were designed and ordered as DNA-oligomers. The inserts were annealed and inserted into the purified PCR product of [http://parts.igem.org/Part:BBa_K1362020 BBa_K1362020] by Golden Gate cloning. | ||
| - | The constructs to express circular xylanase [http://parts.igem.org/Part:BBa_K1362022 (BBa_K1362022)], circular xylanase with His6-tag [http://parts.igem.org/Part:BBa_K1362023 (BBa_K1362023)] and non-circular xylanase with His6-tag (with a Cys->Gly mutation in the C-extein) are based on the [http://parts.igem.org/Part:BBa_K1362000 NpuDnaE intein RFC[i] circularization construct]. For [http://parts.igem.org/Part:BBa_K1362022 BBa_K1362022], the coding sequence of xylanase without start codon, export signal and stop codon was amplified from ''B. subtilis'' by PCR. Overhangs containing RFC[i] E standard overhang, BsaI sites, the coding sequences of the exteins CWE (forward) and RGK, the RFC[i] B standard overhang, BsaI (reverse) were added by primer overhangs. The PCR product was purified and it was inserted into the [http://parts.igem.org/Part:BBa_K1362000 NpuDnaE intein RFC | + | The constructs to express circular xylanase [http://parts.igem.org/Part:BBa_K1362022 (BBa_K1362022)], circular xylanase with His6-tag [http://parts.igem.org/Part:BBa_K1362023 (BBa_K1362023)] and non-circular xylanase with His6-tag (with a Cys->Gly mutation in the C-extein) are based on the [http://parts.igem.org/Part:BBa_K1362000 NpuDnaE intein RFC[i] circularization construct]. For [http://parts.igem.org/Part:BBa_K1362022 BBa_K1362022], the coding sequence of xylanase without start codon, export signal and stop codon was amplified from ''B. subtilis'' by PCR. Overhangs containing RFC[i] E standard overhang, BsaI sites, the coding sequences of the exteins CWE (forward) and RGK, the RFC[i] B standard overhang, BsaI (reverse) were added by primer overhangs. The PCR product was purified and it was inserted into the [http://parts.igem.org/Part:BBa_K1362000 NpuDnaE intein RFC{i} circularization construct] by Golden Gate assembly. [http://parts.igem.org/Part:BBa_K1362023 BBa_K1362023] and the construct to express non-circular xylanase with His6-tag (with a Cys->Gly mutation in the C-extein) were cloned in the same way, except for the additional His6-tags in the primer overhangs. |
==Xylanase Assay== | ==Xylanase Assay== | ||
Revision as of 00:30, 18 October 2014
Contents |
Introduction
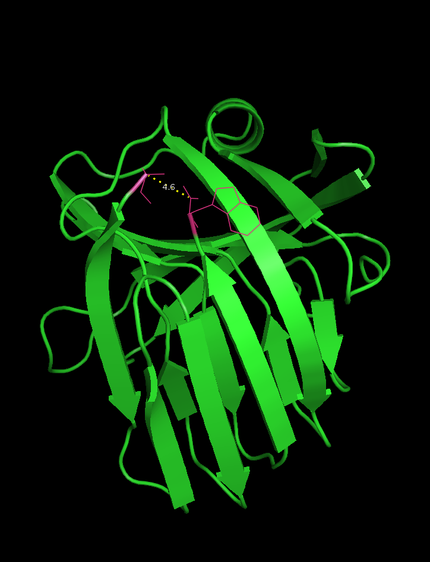
We want to introduce intein-mediated circularization as an alternative approach to confer heat stabilityproteins. We chose the xylanase of B. subtilis which is a promising enzyme to be used in pulp bleaching. Moreover, N- and C-terminus of this xylanase are in close (4.6 Å , see Figure 1) which facilitates circularization. To achieve circularization we used split inteins . This method produces after the splicing reaction a scar of 6 amino acids, which can be bridge the gap between the native termini.
Experimental Procedures
Design and cloning of constructs
To generate a circular, heat-stable xylanase we chose to follow two strategies and circularize not only the native form but also a xylanase including a histidine tag for purification. For circularization we used the split NpuDnaE intein, since it is used as the golden standard for protein splicing. All cloning steps were carried out using our new RFC[i] standard procedure. An overview of all constructs including controls for circularization is shown in the following table:
| Construct Names | Motivation |
| Linear | native xylanase |
| Linear +His | control for “Circular with His6-Tag” construct |
| Linear+His+ Ext | control with similar amino acid composition as the circular His6-tag xylanase (“Circular with His-tag”) |
| Circular | native xylanase with split NpuDnaE split intein |
| Circular + His | native xylanase with split NpuDnaE split intein and His6-tag |
| Circular +His+ destr.Ext. | control with mutation in C-Ext. (Cys-> Gly) |
For the construct to express linear xylanase [http://parts.igem.org/Part:BBa_K1362020 (BBa_K1362020)] the coding sequence of xylanase without start codon and export signal was amplified from B. subtilis by PCR. Overhangs containing an EcoRI site, the RBS [http://parts.igem.org/Part:BBa_B0034 B0034], a start codon (forward) and a SpeI site and a PstI site (reverse) were added by primer overhangs. The PCR product was cut with X and P and ligated into the expression vector [http://parts.igem.org/Part:BBa_K1362093 pSBX1K30]. In order to create the constructs to express linear xylanase with His6-Tag [http://parts.igem.org/Part:BBa_K1362023 BBa_K1362023] and linear xylanase with His6-tag and exteins, primers to linearize [http://parts.igem.org/Part:BBa_K1362020 BBa_K1362020] between the xylanase coding sequence and the stop codon and add BsaI sites at the ends were designed. Inserts coding for the sequence GSHHHHHHSG (His6-Tag) respectively HHHHHHRGKCWE (His6-tag + exteins) with overhangs corresponding to the overhangs caused by BsaI restriction of the vector PCR product were designed and ordered as DNA-oligomers. The inserts were annealed and inserted into the purified PCR product of [http://parts.igem.org/Part:BBa_K1362020 BBa_K1362020] by Golden Gate cloning.
The constructs to express circular xylanase [http://parts.igem.org/Part:BBa_K1362022 (BBa_K1362022)], circular xylanase with His6-tag [http://parts.igem.org/Part:BBa_K1362023 (BBa_K1362023)] and non-circular xylanase with His6-tag (with a Cys->Gly mutation in the C-extein) are based on the [http://parts.igem.org/Part:BBa_K1362000 NpuDnaE intein RFC[i] circularization construct]. For [http://parts.igem.org/Part:BBa_K1362022 BBa_K1362022], the coding sequence of xylanase without start codon, export signal and stop codon was amplified from B. subtilis by PCR. Overhangs containing RFC[i] E standard overhang, BsaI sites, the coding sequences of the exteins CWE (forward) and RGK, the RFC[i] B standard overhang, BsaI (reverse) were added by primer overhangs. The PCR product was purified and it was inserted into the [http://parts.igem.org/Part:BBa_K1362000 NpuDnaE intein RFC{i} circularization construct] by Golden Gate assembly. [http://parts.igem.org/Part:BBa_K1362023 BBa_K1362023] and the construct to express non-circular xylanase with His6-tag (with a Cys->Gly mutation in the C-extein) were cloned in the same way, except for the additional His6-tags in the primer overhangs.
Xylanase Assay
To test if circularization increases heat stability of B. subtilis xylanase we determined xylanase activity after 30min incubation at different temperatures using the EnzChek® Ultra Xylanase Assay Kit. This kit includes a fluorescently labelled hemicellulose substrate. Xylanase-mediated hydrolysis of the substrate results in the unquenching of the attached fluorescent dyes and thus increased fluorescence. Fluorescence intensity over time was measured using the Tecan Infinite M200 reader and xylanase from Thermomyces lanuginosus was used as an initial positive control. Data were analyzed and visualized using a python script similar to the one for the lysozyme assay.
Results
1) Expression of linear and circular Xylanase
To generate a circular, heat-stable xylanase we chose to follow two strategies and circularize not only the native form but also a His6-tagged xylanase. His-tagged xylanase can be easily purified and thus, xylanase activity can be determined more reliably and without interfering molecules that are otherwise present in the lysate. On the other hand, the His-tag might influence thermostability and xylanase activity. Therefore we also included the unmodified xylanase and measured activity directly in bacterial lysate similar to the lysozyme assay.
For circularization we used the split NpuDnaE intein and performed cloning using our new RFC[i] standard procedure.
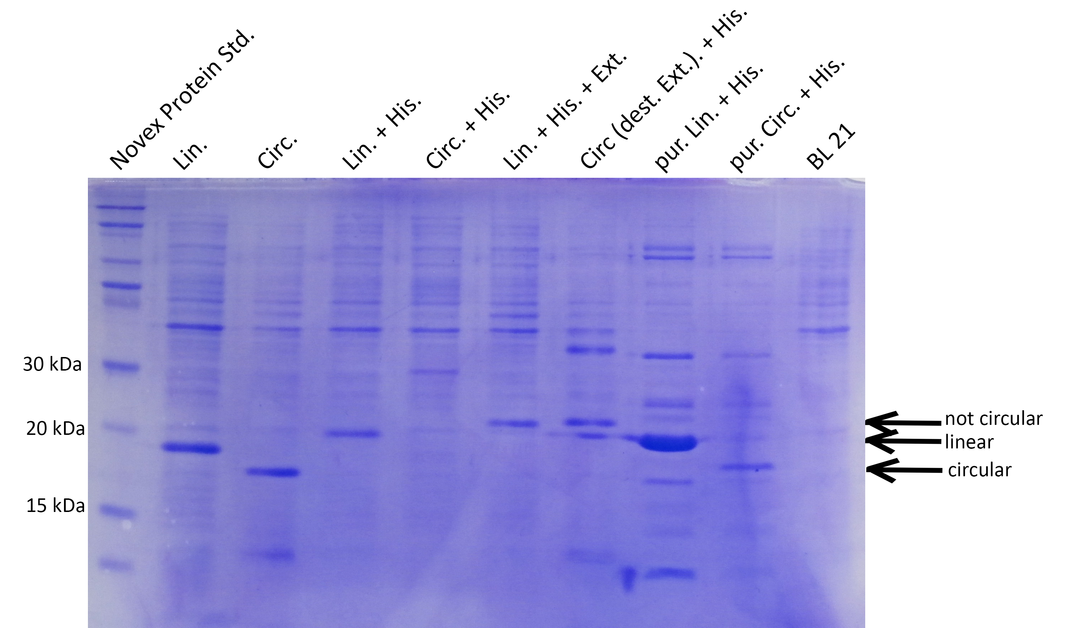
BL21 cultures were induced with IPTG, harvested, lysed and analyzed by SDS-PAGE. Compared to the negative control (BL21), prominent bands at 15-20kDa indicate that all constructs except the circular His-tagged xylanase (Circ.His) are efficiently expressed. Circularization is specific and not achieved with destroyed exteins (no band of circular xylanase at ~18kDa for Circ.destr.Ext+His) (Lin.=linear, Circ.=circular, His.=Histidine tag, Ext.=extein, dest.=destroyed, pur=purified with His-trap column).
Successful cloning and expression of the different linear and circular xylanase variants was verified by a Coomassie stained SDS polyacrylamide gel (see Figure 1). The prominent bands at ~20kDa indicate that linear xylanase with and without His-Tag (Lin., Lin.His., Figure 1) could be efficiently expressed in E.coli BL21. From the circular xylanase constructs the circular variant (Circ.) was produced in satisfying amounts whereas the His-tagged variant was not efficiently expressed (Circ, Circ.His, Figure 1) Nevertheless, the His-tagged circular xylanase could be enriched by purification using a His-Trap column (Circ. (p)+His, Figure 1).
Interestingly, the circular xylanases (Circ, Circ.(p).His) appear to have a lower molecular weight (at ~18kDa) on the gel than their linear counterparts (at ~20kDa) although they include six amino acids of exteins not present in the linear forms. This observation can be explained by the different migration properties of linear and circular molecules during gel electrophoresis. Similar to circular plasmids which migrate faster than their respective linearized versions in agarose gels, circularized proteins such as GFP move faster than their linear counterparts (2). Since the circularization control construct with the destroyed extein (Circ., destr. extein)+His) does not show a band at the same height as the circular xylanases, circularization seems to be specific. Taken together, the circularization control and migration behavior of our circular constructs strongly indicate that we succeeded in circularizing xylanase.
To proof circularization, we cooperated with Dr. Martina Schnölzer and Dr. Uwe Warnken from the mass spectrometry facility of the German Cancer Research center. In the facility, circularized and linear xylanase were extracted from the gel, digested with chymotrypsin and analyzed by ESI mass spectrometry. Unfortunately, we do not have the final results yet but are confident to present them at the jamboree.
2) Linear and circular xylanases show different activities after heat exposure
After the successful expression of linear and circular xylanase we analyzed how circularization influences resistance towards heat. We measured the activity of purified linear and circular xylanase after 30min heat exposure.
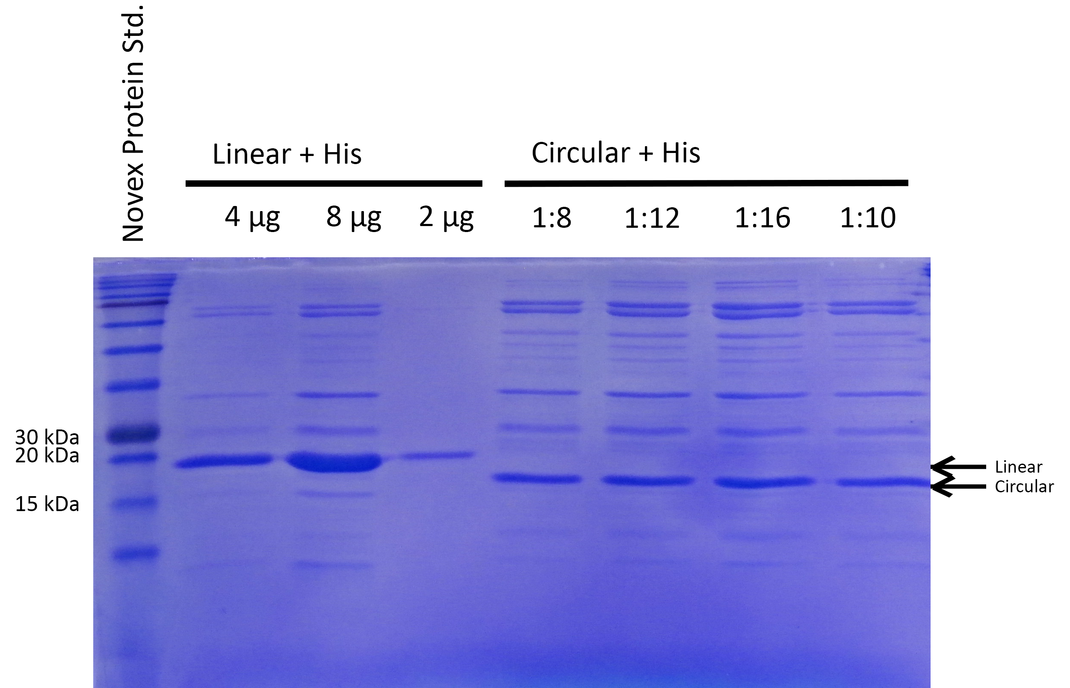
To be able to compare the activity of linear and circular forms, the enzyme amounts in the different samples were estimated by absorption measurement at 280 nm and verified by SDS PAGE (see Figure 2). By determining intensities of the Xylanase bands with ImageJ we estimated a ratio of 1:8 of linear and circular form, respectively.
Our initial results show an inconsistent behavior of purified linear and circular xylanase activity after heat shock at different temperatures. Since we did not observe similar activity at 37°C for circular and linear xylanase, the activity after heat shock cannot be compared. To exclude potential effects of the His-tag, we repeated the activity assay with the unpurified, native linear and circular xylanase. Both, unpurified linear and circular xylanase lose completely their activity over 50 °C which has been previously reported ([http://www.ncbi.nlm.nih.gov/pmc/articles/PMC239434/]). Similar to the His-tag containing xylanases, temperature-dependent activity of the unpurified linear and circular xylanase cannot be compared due to different activity at 37°C.
Linear and circular xylanase were incubated in assay buffer for 30min at the indicated temperatures and added to the hemicellulose substrate. Fluorescence intensities (relative fluorescence units) were measured over 45min
Linear and circular xylanase were incubated in assay buffer for 30min at the indicated temperatures and added to the hemicellulose substrate. Fluorescence intensities (relative fluorescence units) were measured over 160min
References
- Sharma, M. and A. Kumar, Xylanases : An Overview. 2013. 3: p. 1-28.
- Iwai, H., Lingel, a & Pluckthun, a. Cyclic green fluorescent protein produced in vivo using an artificially split PI-PfuI intein from Pyrococcus furiosus. J. Biol. Chem. 276, 16548–54 (2001).
 "
"