Team:Tianjin/Project
From 2014.igem.org
(Difference between revisions)
| (One intermediate revision not shown) | |||
| Line 26: | Line 26: | ||
width:200px; | width:200px; | ||
position:fixed; | position:fixed; | ||
| - | top: | + | top:290px; |
right:10px; | right:10px; | ||
border: medium dashed #C67B14; | border: medium dashed #C67B14; | ||
| Line 479: | Line 479: | ||
<table width="100%" border="3" cellpadding="0" cellspacing="0" bordercolor="#C67B14" bgcolor="#993300"> | <table width="100%" border="3" cellpadding="0" cellspacing="0" bordercolor="#C67B14" bgcolor="#993300"> | ||
<tr> | <tr> | ||
| - | <td bgcolor="#FFED97"><p class="STYLE33"> | + | <td bgcolor="#FFED97"><p class="STYLE33">To achieve the convert from bio-respond to electrical output, we designed the transmitter "transfibre", and the mechanics of which can be summarized as follow: On appearance of the substance of interest, our bacteria will inductively grow a large quantity of modified curli fibers which can adsorb nanogold. These nanowires are highly exclusive in conductivity from the culture, thus, they will conductively bridges two separated electrodes that pre-arranged in the culture and the change of current can be monitored, which present the approximate concentration of the target substance. </p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">Our design work of this bio-electrical bridge can be summarized into following parts. </p> |
| - | <p class="STYLE33"><strong>Part | + | <p class="STYLE33"><strong>Part 1: the controllable expression of the curli fiber</strong> </p> |
| - | <p class="STYLE33">& | + | <p class="STYLE33">1.Background </p> |
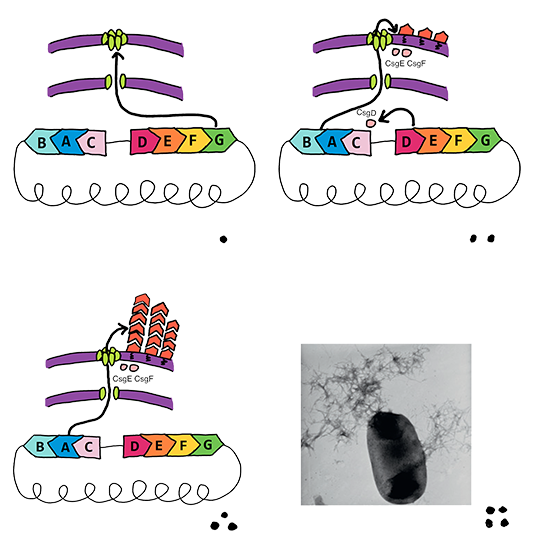
| + | <p class="STYLE33">The delicate curli-assembly system in E. coli K-12 is revealed lately and chose for our "transfibre" prototype. Curli fibers, that play key role in establishment and persistence of biofilm communities, are coded by the Csg gene cluster where seven genes(CsgABCDEFG) located, and their function are listed here: <br /> | ||
| + | CsgA: At the cell surface, CsgA, The main subunit, alone soluble in vivo, that can be nucleated into an amyloid fiber by CsgB. <br /> | ||
| + | CsgB: Cell-surface-associated nucleation agent. <br /> | ||
| + | CsgC: CsgC is less well understood and it was predicted to have oxidoreductase activity. Within the periplasm, CsgC may regulate CsgG outer membrane assembly and pore activity through modification of C230 in CsgG. <br /> | ||
| + | CsgE: a small periplasmic protein, is proposed to direct CsgA to the CsgG secretion complex and helps mediate secretion specificity via an N-terminal sequence in the mature CsgA. CsgE also inhibits CsgA polymerization in vitro, and hence CsgE could be considered a CsgA-specific chaperone. <br /> | ||
| + | CsgF: CsgF is a membrane-associated protein, required for CsgB surface exposure, and is crucial for efficient CsgA polymerization. <br /> | ||
| + | CsgG: Once in the periplasm, the lipoprotein CsgG is transported by the LOL (lipoprotein outer-membrane localization) transport system to the outer membrane where it oligomerizes into a pore-like structure that is required for secretion of CsgA and CsgB into the extracellular milieu. </p> | ||
<table width="100%" border="0" cellspacing="0" cellpadding="0"> | <table width="100%" border="0" cellspacing="0" cellpadding="0"> | ||
<tr> | <tr> | ||
| - | <td bgcolor="#FFED97"><span class="STYLE33"><a href="https://static.igem.org/mediawiki/2014/ | + | <td bgcolor="#FFED97"><span class="STYLE33"><a href="https://static.igem.org/mediawiki/2014/1/19/Df.png"><img src="https://static.igem.org/mediawiki/2014/1/19/Df.png" width="557" height="545" border="0" /></a></span></td> |
| + | </tr> | ||
| + | <tr> | ||
| + | <td bgcolor="#FFED97"><span class="STYLE33">Fig 1. This figure demonstrates the cooperation of curli-associated genes when bacteria respond to external environment change.</span></td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | <p class="STYLE33"> | + | <p class="STYLE33">For the reason that curli fiber mostly acts an emergency mechanism that could enhance bacterial survival in extreme conditions such as low osmolality, low growth temperature (<32°C) and hydroponic, the regulation system attached strictly limited the expression of two curli operons under normal circumstances that bio-sensing processes operating. </p> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<table width="100%" border="0" cellspacing="0" cellpadding="0"> | <table width="100%" border="0" cellspacing="0" cellpadding="0"> | ||
<tr> | <tr> | ||
| - | <td bgcolor="#FFED97"><span class="STYLE33"><a href="https://static.igem.org/mediawiki/2014/ | + | <td bgcolor="#FFED97"><span class="STYLE33"><a href="https://static.igem.org/mediawiki/2014/5/59/11.png"><img src="https://static.igem.org/mediawiki/2014/5/59/11.png" width="760" height="520" border="0" /></a></td> |
| + | </tr> | ||
| + | <tr> | ||
| + | <td bgcolor="#FFED97"><span class="STYLE33">Fig 2. Genes that associated with curli fiber assembly are located on two operons in E coli. K-12 genome. They are under strict control and low level of expression at normal cultivation conditions. </span></td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | <p class="STYLE33"> | + | <p class="STYLE33">2. The design idea <br /> |
| - | + | </p> | |
| - | + | <p class="STYLE33">According to the synthetic properties of curli fiber, CsgA interacts with molecular chaperones CsgE and maintains it in a soluble state in vivo, preventing CsgA from aggregation without CsgB protein. Thus, controllable expression of CsgB can provide nucleation for CsgA rapid aggression and recruits CsgA monomers that pre-secreted in culture into fibers, which meets the request of controllable expression and immediate respond for biosensor output platform. <br /> | |
| - | <p class="STYLE33"> | + | Following this strategy, we devise an inducible expression system of CsgB and a constitutive expression system of the other Csg-genes (excluding CsgD). Referring to the original curli assembly device, CsgA, the major subunit of curli fiber, and CsgC are constructed in a high copy plasmid (pSB1C3) and constitutive expression by a relatively strong promoter, while CsgEFG using a weaker promoter and docked in pSB3K3. <br /> |
| - | + | </p> | |
| - | <p class="STYLE33"> | + | <p class="STYLE33">To obtain target genes from Csg operons, method PCR was applied on the genome of DH5alpha strains (The original genes of CsgA and CsgE has SpeI and PstI restriction enzyme cutting sites. They have been substituted by synonym codon according to codon usage bias). Then, construct expression vector of these genes to transform E coli. BL21 and transT-1. (In BL21, CsgA and CsgB genes has been knocked out by λ-red recombinase system to eliminate disturb.) </p> |
<table width="100%" border="0" cellspacing="0" cellpadding="0"> | <table width="100%" border="0" cellspacing="0" cellpadding="0"> | ||
<tr> | <tr> | ||
| - | <td bgcolor="#FFED97"><span class="STYLE33"><a href="https://static.igem.org/mediawiki/2014/ | + | <td bgcolor="#FFED97"><span class="STYLE33"><a href="https://static.igem.org/mediawiki/2014/7/7c/完成版.png"><img src="https://static.igem.org/mediawiki/2014/7/7c/完成版.png" width="760" height="380" border="0" /></a></span></td> |
| + | </tr> | ||
| + | <tr> | ||
| + | <td bgcolor="#FFED97"><span class="STYLE33">Fig.3 The concept of bio-electrical transmit. The modified CsgAs are aggregate on the inductive expression of CsgB and bridges the circuit.</span></td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | <p class="STYLE33"> | + | <p class="STYLE33">Two promoters, T7 promoter and Pbad promoter, are selected whose inducers, IPTG and L-arabinose respectively, are chosen to simulate the target inducing process. Meanwhile, for CsgA and CsgE, F, G, we also choose three promoters with different strength--- P1 (BBa_J23100), P2 (BBa_J23104) and P3(BBa_J23110) to screen out a better gene expression combination through this method. Thus, six parts are constructed with different levels of CsgA, C expression to work out the best fiber polymerization. </p> |
| - | + | ||
<table width="100%" border="0" cellspacing="0" cellpadding="0"> | <table width="100%" border="0" cellspacing="0" cellpadding="0"> | ||
<tr> | <tr> | ||
| - | <td bgcolor="#FFED97"><table border="1" | + | <td bgcolor="#FFED97"><table border="1" cellspacing="0" cellpadding="0" width="559"> |
<tr> | <tr> | ||
| - | <td width=" | + | <td width="559" colspan="2" valign="top"><p class="STYLE33">Biobricks for Promoter Strength Trail</p></td> |
| - | + | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td width=" | + | <td width="253" valign="top"><p class="STYLE33">T7-CsgB-P1-CsgA, C</p></td> |
| - | <td width=" | + | <td width="307" valign="top"><p class="STYLE33">Pbad-CsgB-P1-CsgA, C</p></td> |
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="253" valign="top"><p class="STYLE33">T7-CsgB-P2-CsgA, C</p></td> | ||
| + | <td width="307" valign="top"><p class="STYLE33">Pbad-CsgB-P1-CsgA, C</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="253" valign="top"><p class="STYLE33">P2-CsgE, F, G</p></td> | ||
| + | <td width="307" valign="top"><p class="STYLE33">P3-CsgE, F, G</p></td> | ||
</tr> | </tr> | ||
</table></td> | </table></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td bgcolor="#FFED97"><span class="STYLE33">Table 1. Six parts with distinguished strength of constitutive promoters were constructed. And combinations of them were tested. </span></td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | <p class="STYLE33"> | + | <p class="STYLE33">In addition, we truncate 19 amino acids from CsgB molecule’s C-terminus and get protein CsgBtrunc. It has been tested to be not outer membrane-associated, but secreted away. Soluble CsgBtrunc can also assemble into fibers that bound to the amyloid-specific dyes Congo red, and CsgBtrunc are able to seed soluble CsgA polymerization in vitro. Although it displayed only modest nucleator activity in vivo, considering that our modules could secrete much higher concentration of CsgA, it could acts relatively good nucleating capacity. </p> |
| - | + | <p align="left" class="STYLE33">The establishment of CsgBtrunc modules resembles with CsgB. The difference is that, after the addition of inducers, the curli fiber will grow suspendingly in liquid, not anchored on the outer membrane of E.coli. </p> | |
| - | <p class="STYLE33"> | + | |
<table width="100%" border="0" cellspacing="0" cellpadding="0"> | <table width="100%" border="0" cellspacing="0" cellpadding="0"> | ||
<tr> | <tr> | ||
| - | <td bgcolor="#FFED97"><a href="https://static.igem.org/mediawiki/2014/ | + | <td bgcolor="#FFED97"><span class="STYLE33"><strong><a href="https://static.igem.org/mediawiki/2014/5/52/已完成2.png"><img src="https://static.igem.org/mediawiki/2014/5/52/已完成2.png" width="480" height="360" border="0" /></a></strong></span></td> |
| + | </tr> | ||
| + | <tr> | ||
| + | <td bgcolor="#FFED97"><span class="STYLE33">Fig 4. This figure shows the concept of CsgBtrunc expression and off-cell nucleating on CsgA. <br /> | ||
| + | 3. An alternative expression strategy</span></td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | <p | + | <p align="left" class="STYLE33">We can transform CsgA and CsgB into separate bacteria and put them in same culture. In this mode, one kind of E. coli acts as ‘material background’ that provide constitutively expressed CsgBtrunc, and another acts as ‘inducible nucleator’ which CsgA is under induction. Detection acting at a distance could be accomplished by this method. However, this method was not accomplished and we don’t know its advantages clearly. </p> |
| - | + | <p class="STYLE33"><strong>Part 2: The method of electric conduction</strong> </p> | |
| - | + | <p class="STYLE33">Our curli fiber is supposed to adsorb nanogold, which is conductive and can act as wires when linked to each other. However, the wild type CsgA monomer is unable to adsorb nanogold. Thus, some measures should be taken to functionize it. <br /> | |
| - | <p class="STYLE33"><strong>Part | + | According to literature research, we designed three methods to achieve this goal: inserting two histidine tags (one before the first repeat domain and one after the last repeat domain in CsgA), adding a cysteine tag and inserting an amino acid sequence of pentapeptide. The principle of these three methods is the same: adding ligands like sulfhydryl that have good affinity with gold to gain CsgA ability of absorbing nanogold. <br /> |
| - | <p class="STYLE33"> | + | As for the first method, CsgA-His is constitutive expression whereas CsgB is under the control of Pbad promoter. Once we adding the inducer, CsgA would quickly aggregates and thus curli fiber with his tags would generate immediately and absorbing Au-NTA-Ni particle. We want to test the affinity between Meanwhile, the CsgA-His can be used for CsgA protein detection and purification. For the second method that adding a cysteine tag, the integral mechanism of generating the curli fibre is same as above, but the sulfhydryl of cysteine tags can link to the nanogold particles itself because the monodisperse spherical gold nanoparticles are arranged to chained structure due to the effect of these molecules. Besides, the citrate layer on the metal surface is replaced by cysteine leading to a formation of organic double layer structure, which enhances the absorption. As for the third one, the composition of the pentapeptide is Leu-Pro-Phe-Phe-Asp-OH (LPFFD-OH), the existence of N-donors containing side-chains will help a lot in the attachment or binding to the surface of gold nanoparticles, though the detailed mechanisms of the binding of the pentapeptide was not fully resolved. </p> |
| - | + | ||
| - | + | ||
<table width="100%" border="0" cellspacing="0" cellpadding="0"> | <table width="100%" border="0" cellspacing="0" cellpadding="0"> | ||
<tr> | <tr> | ||
| - | <td bgcolor="#FFED97"><span class="STYLE33">< | + | <td bgcolor="#FFED97"><span class="STYLE33"><img width="760" height="560" src="https://static.igem.org/mediawiki/2014/a/ac/已完成.png" /></span></td> |
| + | </tr> | ||
| + | <tr> | ||
| + | <td bgcolor="#FFED97"><span class="STYLE33">Fig 5. The modified CsgA that obtains the ability of absorbing nanogold will firm conductive wires between electrodes on the respond of inducer.</span></td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | <p class="STYLE33"> | + | <p class="STYLE33">In all three methods, the primary concept is to produce conductible curli fiber and accomplish the bio-signal transformation. We have already known the modified curli fiber were produced by a certain kind of inducer (arabinose and IPTG were used in our experiments) and then attached to AuNPs. Since we assumed that the intensity of the induction could be reflected by the quantity of the AuNPs-linked curli, the concentration of the inducer could thus determine the resistance and consequent current of the whole circuits. In this way, we transformed the bio-signal into the electric signal. Furthermore, as the system is highly modularized, we can substitute the promoter of other inducers so that we could extend this system to detect sorts of subsidence apart from arabinose we used. </p> |
| - | + | <p class="STYLE33"><strong>Part 3: the upgrade of idea-logic gate.</strong> </p> | |
| - | <p class="STYLE33"><strong>Part | + | <p class="STYLE33">As mentioned, CsgA will aggregate into curli fibers with the seduction by seducer (like arabinose). In order to make the whole system a contenting device to characterize the concentration of seducer, a NOT gate is added to sabotage the nanowire. Thus, the current will decline and the change of concentration can still follow a kind of function, which can be determined by experiments. <br /> |
| - | <p class="STYLE33"> | + | AuNPs-based nanowire may be digested by protease or tear into pieces under the influence of physical forces. Because of the endurance of CsgA to proteases, we design a forces-destroy pathway. <br /> |
| - | + | CheY and CheZ are antagonist in a system regulating the motion and the resulting chemotaxis of E.coli: Phosphorylated CheY motivates the rolling of the bacterium, and it is damped by CheZ through prompting the dephosphorylation. <br /> | |
| - | + | We construct two plasmids. One is BBa_K1361000, which has the stronger promoter P1, and the other has the weaker promoter P2. CI is a repressor gene that can restrain the expression of CheZ, and Plam acts as the promoter to start the gene of expression of cheZ. If the concentration of the inducer increases, the Pbad promotes the gene expression of CsgB as well as the repressor CI. So the effect of inhibition of CheZ increases as the curli fiber grows, because the repressor CI can bind to the Plam and inhibit the expression of CheZ. Thus, CheZ gene is depressed. If the concentration of the inducer decreases, the concentration of the repressor decreases. So the promoter Plam could start the expression of CheZ in some degree. The CheZ gene expresses and it can and, afterwards, motivate the motion of flagellum, that naturally destroyed curli fiber. So the intensity of the electric current will descend. </p> | |
| - | + | ||
| - | + | ||
| - | + | ||
<table width="100%" border="0" cellspacing="0" cellpadding="0"> | <table width="100%" border="0" cellspacing="0" cellpadding="0"> | ||
<tr> | <tr> | ||
| - | <td bgcolor="#FFED97"><span class="STYLE33"><a href="https://static.igem.org/mediawiki/2014/ | + | <td bgcolor="#FFED97"><span class="STYLE33"><a href="https://static.igem.org/mediawiki/2014/7/7a/9.png"><img src="https://static.igem.org/mediawiki/2014/7/7a/9.png" width="796" height="579" border="0" /></a></span></td> |
| + | </tr> | ||
| + | <tr> | ||
| + | <td bgcolor="#FFED97"><span class="STYLE33">Fig 6. The mechanic of CheZ-Curli interaction is demonstrated in the figure, hypothesizing that the increasing motility of cells will destruct the curli fiber. </span></td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | <p class="STYLE33"> | + | <p class="STYLE33">Because our conductive biofilm build a bridge between bacteria and electricity, our E.coli is expected to operate machine. As the concentration of inducer increases, the circuit can be turned on, and it can switch almost any electrical devices and machines with or without the help of signal amplifier. It, which may extend the application of transfibre. </p> |
| - | + | ||
<table width="100%" border="0" cellspacing="0" cellpadding="0"> | <table width="100%" border="0" cellspacing="0" cellpadding="0"> | ||
<tr> | <tr> | ||
| - | <td bgcolor="#FFED97"><span class="STYLE33"><a href="https://static.igem.org/mediawiki/2014/ | + | <td bgcolor="#FFED97"><span class="STYLE33"><a href="https://static.igem.org/mediawiki/2014/7/7f/已完成_(2).png"><img src="https://static.igem.org/mediawiki/2014/7/7f/%E5%B7%B2%E5%AE%8C%E6%88%90_%282%29.png" width="442" height="327" border="0" /></a></span></td> |
| + | </tr> | ||
| + | <tr> | ||
| + | <td bgcolor="#FFED97"><span class="STYLE33">Fig 7. The schematic diagram of coordination between bio-device and machines.</span></td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | + | <p class="STYLE33"><strong>Part 4: The detection device</strong> </p> | |
| - | <p class="STYLE33"><strong>Part | + | <p class="STYLE33">Our living cell system can be used to create environmentally switchable conductive biofilms. We hypothesized that IPTG-or-L-arabinose-inducible production of CsgB monomers by our cells would generate extracellular amyloid fibrils that organize AuNPs into chains, and form a conductive biofilm network. <br /> |
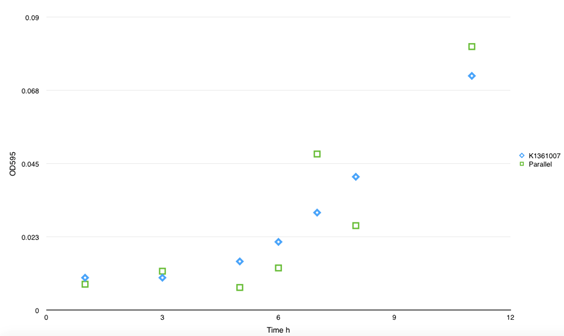
| - | <p class="STYLE33"> | + | Engineered biofilms were grown on interdigitated electrodes deposited on glass slides, with our E.coli cultured in the presence of AuNPs and in the presence or absence of IPTG or L-arabinose inducer. </p> |
| - | + | ||
<table width="100%" border="0" cellspacing="0" cellpadding="0"> | <table width="100%" border="0" cellspacing="0" cellpadding="0"> | ||
<tr> | <tr> | ||
| - | <td bgcolor="#FFED97"><span class="STYLE33">< | + | <td bordercolor="#FFED97" bgcolor="#FFED97"><span class="STYLE33"><img width="760" height="200" src="https://static.igem.org/mediawiki/2014/a/a9/7676.png" /></span></td> |
| + | </tr> | ||
| + | <tr> | ||
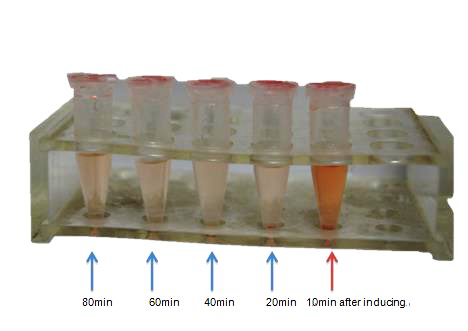
| + | <td bgcolor="#FFED97"><span class="STYLE33">Fig 8. The picture shows the making process from test tube to our detection petri dish.<br /> | ||
| + | We designed the petri dish by ourselves. For the convenience of depositing electrodes, we composed a detachable culture dish. Firstly, we cut the test tube into a 3cm-long section without bottom. Secondly, we cut another test tube with longer diameter into a 4cm-long section with a bottom. The agar is utilized to attach the 3cm-long section to a glass slide to make the petri dish, and the 4cm-long section acts as a cover to ensure sterility. </span></td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | |||
| - | |||
<table width="100%" border="0" cellspacing="0" cellpadding="0"> | <table width="100%" border="0" cellspacing="0" cellpadding="0"> | ||
<tr> | <tr> | ||
| - | <td bgcolor="#FFED97"><span class="STYLE33"><a href="https://static.igem.org/mediawiki/2014/ | + | <td bgcolor="#FFED97"><span class="STYLE33"><a href="https://static.igem.org/mediawiki/2014/8/80/仪器.JPG"><img src="https://static.igem.org/mediawiki/2014/8/80/仪器.JPG" width="708" height="497" border="0" /></a></span></td> |
| + | </tr> | ||
| + | <tr> | ||
| + | <td bgcolor="#FFED97"><span class="STYLE33">Fig 9. The real object of our self-made petri dish.</span></td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | <p class="STYLE33"> | + | <p class="STYLE33">The fabrication of electrodes resembles the method of Synthesis and patterning of tunable multiscale materials with engineered cells. Interdigitated electrodes for measuring biofilm conductance were created by sputtering gold through shadowing masks onto glass slides. </p> |
| - | + | <p class="STYLE33">Our circuit is designed by Yuanchi Ma at College of Precision Instrument& Opto-Electronics Engineering. </p> | |
<table width="100%" border="0" cellspacing="0" cellpadding="0"> | <table width="100%" border="0" cellspacing="0" cellpadding="0"> | ||
<tr> | <tr> | ||
| - | <td bgcolor="#FFED97"><span class="STYLE33"><a href="https://static.igem.org/mediawiki/2014/ | + | <td bgcolor="#FFED97"><span class="STYLE33"><a href="https://static.igem.org/mediawiki/2014/9/99/DSC00001.png"><img src="https://static.igem.org/mediawiki/2014/9/99/DSC00001.png" width="560" height="373" border="0" /></a></span></td> |
| + | </tr> | ||
| + | <tr> | ||
| + | <td bgcolor="#FFED97"> </td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | + | <p class="STYLE33"><strong>Part 5: Stabilized matrix sensor for MMP-7</strong> </p> | |
| - | + | <p class="STYLE33">The qualitative or quantitative detection of proteases can be achieved by various methods such as mass spectrum, two-dimensional gel electrophoresis and antigen-antibody complex reaction. However, they are usually time-consumed, demanding and the data is difficult to be stored and processed. Our project can transfer bio-signal to electrical signal, and as a result, we try to make it a convenient device to detect proteases without those aforementioned drawbacks. </p> | |
| - | <p class="STYLE33"><strong>Part | + | <p class="STYLE33">MMP-7 is the abbreviation of matrix metalloproteinase-7, which belongs to the family of MMPs (a family of extracellular zinc proteases that have long been associated with tumor invasion and metastasis and thus may serve as important cancer biomarkers). It has be proved that a helix peptide (Arg-Pro-Leu-Ala-Leu-Trp-Arg-Ser) is the substrate of MMP-7. </p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">The substrate peptide of MMP-7 and binding-site peptide of nano-gold particles (AuNPs) are modified with CsgA. These two modules added to the C-terminal of the wild type CsgA will not seriously interrupt the normal aggregation of CsgA. Consequently, we get redesigned curli fibers with the capacity to detect the MMP-7: When no MMP-7 exists in the system with applied voltage, the most electrons flows through the nanowire composed by AuNPs binding to the modified CsgAs; however, the resistance of the fibers will skyrocket for the detrimental enzymolysis of the substrate peptide by MMP-7, which result in the wreck of nanowire. Based on this blueprint, the existence of MMP-7 can be detected and the concentration can be calculated by the concentration function of current. </p> |
| - | <p class="STYLE33"> | + | |
| - | <p class="STYLE33"> | + | |
<table width="100%" border="0" cellspacing="0" cellpadding="0"> | <table width="100%" border="0" cellspacing="0" cellpadding="0"> | ||
<tr> | <tr> | ||
| - | <td><span class="STYLE33"><a href="https://static.igem.org/mediawiki/2014/ | + | <td bgcolor="#FFED97"><span class="STYLE33"><a name="_GoBack" id="_GoBack"><a href="https://static.igem.org/mediawiki/2014/b/b1/MMP-7插图,已完成.png"><img src="https://static.igem.org/mediawiki/2014/b/b1/MMP-7插图,已完成.png" width="790" height="359" border="0" /></a></td> |
</tr> | </tr> | ||
| - | + | <tr> | |
| - | + | <td bgcolor="#FFED97"><span class="STYLE33">Fig 10. Detection of MMP-7</span></td> | |
| - | <p class="STYLE33">Reference:</ | + | </tr> |
| - | + | </table> | |
| - | + | <p class="STYLE33">Reference: <br /> | |
| - | + | [1] Dobson, C.M. (2005) Structural biology: prying into prions. Nature 435,747–749 <br /> | |
| - | + | [2] Collinson, S.K. et al. (1999) Structural predictions of AgfA, the insoluble fimbrial subunit of Salmonella thin aggregative fimbriae. J. Mol. Biol. 290, 741–756 <br /> | |
| - | + | [3] Shewmaker, F. et al. (2009) The functional curli amyloid is not based on in-register parallel beta-sheet structure. J. Biol. Chem. 284, 25065–25076 <br /> | |
| - | + | [4] Wang, X. et al. (2007) In vitro polymerization of a functional scherichia coli amyloid protein. J. Biol. Chem. 282, 3713–3719 <br /> | |
| - | + | [5] Barnhart, M.M. and Chapman, M.R. (2006) Curli biogenesis and function. Annu. Rev. Microbiol. 60, 131–147 <br /> | |
| - | + | [6] Wang, X. and Chapman, M.R. (2008) Curli provide the template for understanding controlled amyloid propagation. Prion 2, 57–60 <br /> | |
| - | + | [7] Stathopoulos, C. et al. (2000) Secretion of virulence determinants by the general secretory pathway in Gram-negative pathogens: an evolving story. Microbes Infect. 2, 1061–1072 <br /> | |
| - | + | [8] Taylor, J.D. et al. (2011) Atomic resolution insights into curli fiber biogenesis. Structure 19, 1307–1316 <br /> | |
| - | + | [9] Nenninger, A.A. et al. (2011) CsgE is a curli secretion specificity factor that prevents amyloid fibre aggregation. Mol. Microbiol. 81, 486–499 <br /> | |
| - | + | [10] Narita, S. et al. (2004) Lipoprotein trafficking in Escherichia coli. Arch. Microbiol. 182, 1–6 <br /> | |
| - | + | [11] Epstein, E.A. et al. (2009) Spatial clustering of the curlin secretion lipoprotein requires curli fiber assembly. J. Bacteriol. 191, 608–615 <br /> | |
| - | + | [12] Robinson, L.S. et al. (2006) Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol. Microbiol. 59, 870–881. <br /> | |
| - | + | [13] Wang, X. et al. (2010) Gatekeeper residues in the major curlin subunit modulate bacterial amyloid fiber biogenesis. Proc. Natl. Acad. Sci.U.S.A. 107, 163–168 <br /> | |
| - | + | [14] Majzik, A. et al. (2010). Functionalization of gold nanoparticles with amino acid, β-amyloid peptides and fragment. Colloids and Surfaces B: Biointerfaces, 81(1), 235-241. <br /> | |
| - | + | [15] Liu G, Wang J, Wunschel DS, Lin Y (2006) Electrochemical proteolytic beacon for detection of matrix metalloproteinase activities. J Am Chem Soc 128(38):12382–12383. doi:10.1021/ja0626638 <br /> | |
| - | + | [16] Hammer N D, Schmidt J C, Chapman M R. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization[J]. Proceedings of the National Academy of Sciences, 2007, 104(30): 12494-12499. <br /> | |
| + | [17] Chen A Y, Deng Z, Billings A N, et al. Synthesis and patterning of tunable multiscale materials with engineered cells[J]. Nature materials, 2014. </p> | ||
| + | <span class="STYLE33">[18] Scheibel T, Parthasarathy R, Sawicki G, et al. Conducting nanowires built by controlled self-assembly of amyloid fibers and selective metal deposition[J]. Proceedings of the National Academy of Sciences, 2003, 100</span></td> | ||
</tr> | </tr> | ||
</table></td> | </table></td> | ||
Latest revision as of 21:34, 17 October 2014
|
|
Welcome to Team Tianjin! |
 |
|
 "
"