Team:Valencia UPV/Project/modules/problem
From 2014.igem.org
| Line 12: | Line 12: | ||
<p>In order to engineer the insect sexual pheromone pathway in our Sexy plant, we had to isolate four genes from different organisms: a desaturase (AtrΔ11), a reductase (HarFAR), an acetyltransferase (EaDAcT) and finally an alcohol oxidase (FAO). As they were coming from very different and not easily accessible organisms (two moths and a plant from Asia), the coding sequences (CDS) of the first three enzymes were obtained by gene synthesis (Integrated DNA Technologies, IDT) after codon usage optimization for N. benthamiana. As for the fourth one (FAO) we tried to amplify it from the genomic DNA of the yeast Candida tropicalis, with no successful results. Nevertheless, the three synthetic genes were sufficient to produce at least two of our target pheromones, the alcohol (Z)-11-hexadecen-1-ol and the acetate (Z)-11-hexadecenyl acetate.</p><br/> | <p>In order to engineer the insect sexual pheromone pathway in our Sexy plant, we had to isolate four genes from different organisms: a desaturase (AtrΔ11), a reductase (HarFAR), an acetyltransferase (EaDAcT) and finally an alcohol oxidase (FAO). As they were coming from very different and not easily accessible organisms (two moths and a plant from Asia), the coding sequences (CDS) of the first three enzymes were obtained by gene synthesis (Integrated DNA Technologies, IDT) after codon usage optimization for N. benthamiana. As for the fourth one (FAO) we tried to amplify it from the genomic DNA of the yeast Candida tropicalis, with no successful results. Nevertheless, the three synthetic genes were sufficient to produce at least two of our target pheromones, the alcohol (Z)-11-hexadecen-1-ol and the acetate (Z)-11-hexadecenyl acetate.</p><br/> | ||
| - | <br/><p style="text-align: right; font-style: italic; font-size: 0.8em; width: | + | <br/><p style="text-align: right; font-style: italic; font-size: 0.8em; width: 850px;"><span class="black-bold">Figure 1</span>. Engineered pheromone production pathway.</p></div><br/> |
<p>All three DNA sequences were domesticated; that is, standardized as GoldenBraid parts and cloned in the pUPD plasmid. Subsequently, each CDS was assembled with the strong constitutive Cauliflower mosaic virus promoter (P35S) and its terminator (T35S) respectively, in a multipartite assembly reaction. P35S is a strong plant constitutive promoter with high expression levels. As result of these assemblies, we obtained three Transcriptional units (TU) ready for plant transformation.</p><br/> | <p>All three DNA sequences were domesticated; that is, standardized as GoldenBraid parts and cloned in the pUPD plasmid. Subsequently, each CDS was assembled with the strong constitutive Cauliflower mosaic virus promoter (P35S) and its terminator (T35S) respectively, in a multipartite assembly reaction. P35S is a strong plant constitutive promoter with high expression levels. As result of these assemblies, we obtained three Transcriptional units (TU) ready for plant transformation.</p><br/> | ||
Revision as of 16:04, 17 October 2014
Project > Modules > Problem

In order to engineer the insect sexual pheromone pathway in our Sexy plant, we had to isolate four genes from different organisms: a desaturase (AtrΔ11), a reductase (HarFAR), an acetyltransferase (EaDAcT) and finally an alcohol oxidase (FAO). As they were coming from very different and not easily accessible organisms (two moths and a plant from Asia), the coding sequences (CDS) of the first three enzymes were obtained by gene synthesis (Integrated DNA Technologies, IDT) after codon usage optimization for N. benthamiana. As for the fourth one (FAO) we tried to amplify it from the genomic DNA of the yeast Candida tropicalis, with no successful results. Nevertheless, the three synthetic genes were sufficient to produce at least two of our target pheromones, the alcohol (Z)-11-hexadecen-1-ol and the acetate (Z)-11-hexadecenyl acetate.
Figure 1. Engineered pheromone production pathway.
All three DNA sequences were domesticated; that is, standardized as GoldenBraid parts and cloned in the pUPD plasmid. Subsequently, each CDS was assembled with the strong constitutive Cauliflower mosaic virus promoter (P35S) and its terminator (T35S) respectively, in a multipartite assembly reaction. P35S is a strong plant constitutive promoter with high expression levels. As result of these assemblies, we obtained three Transcriptional units (TU) ready for plant transformation.
To maximize the flow through of the pathway, we wanted to make sure that all three genes were co-delivered simultaneously. This is to ensure that each transformed cell receives a complete set of genes and that the expression of all three enzymes is balanced and coordinated. Co-delivery is achieved by creating a multigenic construct, where all three genes are assembled in a single plasmid.
We used the GoldenBraid assembly system to create the multigene assembly. After two binary reactions, the 3-genes construct was obtained. This construct was then transformed, by agroinfiltration, into N. benthamiana plants for pheromone production.
We used GoldenBraid 2.0 for all our cloning reactions. GoldenBraid and BioBrick parts are not directly exchangeable; however, we adapted the coding sequences of the three biosynthetic genes to BioBrick standards using a GoldenBraid-Biobricks translator developed by the NRP-UEA-Norwich team . . These BioBricks have been submitted to the Parts Registry.
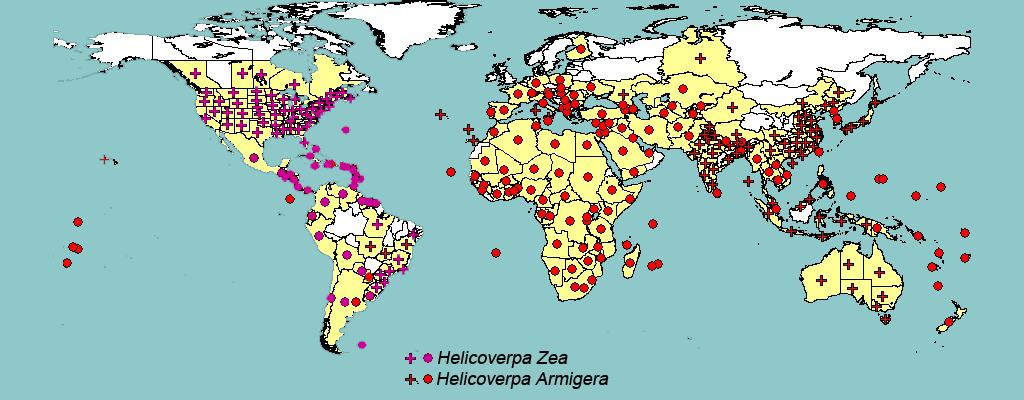
Moth pests are particularly damaging and they affect high number of crops all over the world. It has been reported that H. armigera larvae attack more than 60 species of cultivated and wold host plants, including Asteraceae, Fabaceae, Malvaceae, Poaceae and Solanaceae familie, i.e. affecting the main commercial crops such as cotton, maize, legumes, tomato and ornamental plants (Fitt 1989, EPPO 2010, CABI 2013).
Figure 2. Armigera larvae and dagmaged leaf.
Figure 3. World distribution of H. Armigera and H. Zea pests.
Traditional methods to fight them, based on chemical pesticides, may cause damage in the environment or even in the consumer. Thus, pesticides are becoming less and less socially accepted. In any case, they are not the best solution. Insects tend to become resistant, so the dose needs to increased, leading to further toxicity. Moreover, broad-spectrum pesticides affect other insects which cause no problem as well.
An alternative pest control strategy should be used fight these pests. To this end, the use of sexual pheromones is becoming popular for traps and also for mating disruption strategies. Female insects produce a pheromone trace which allows the male to find them. Once together, they mate and the female lays eggs on the plant. Then the larvae are born and eat the crops. However, if we load the air with pheromones so that its concentration passes a certain threshold, the male won't be able to follow female´s trace anymore and the larvae will never be born. The same principle is used by pheromone traps.
Nowadays the only way to obtain pheromones is by chemical synthesis. Chemical synthesis requires very skilled staff and special installations to get rid of the toxic residues, what leads to a more expensive end product. Alternative cheaper, environmental-friendly production systems are, thus, of great interest.
Our goal with SexyPlant is to create a plant able to produce the pheromones, able to release them into the environment, and completely safe both for humans and environment. Achieving our goal implies solving problems related to:
- Expressing pheromone biosynthesis in the plant by introducing the appropriate pathway
- Achieving pheromone release
- Switching to release the pheromone to the environment just when desired
- Making the plant biosafe
- Analysing pheromone productivity, and optimal distribution of SexyPlant when intercropped with the affected plant species.
 "
"