Team:KAIT Japan/Results
From 2014.igem.org
(→GFP) |
|||
| (47 intermediate revisions not shown) | |||
| Line 53: | Line 53: | ||
| style="width:30% style="vertical-align:top" | | | style="width:30% style="vertical-align:top" | | ||
| - | < | + | =<font size="7">Results</font>= |
<br> | <br> | ||
| - | + | <H1>GFP+HlyA(BBa_K1414000) and HlyA+GFP(BBA_K1414001)</H1> | |
| - | + | ||
{| align="left" style="width:100%; margin-right:0.2em; border-color:#99F; border-style:dotted; border-collapse:collapse; background-color:#FFF;" | {| align="left" style="width:100%; margin-right:0.2em; border-color:#99F; border-style:dotted; border-collapse:collapse; background-color:#FFF;" | ||
| style="width:150px" | | | style="width:150px" | | ||
[[File:GFP.jpg]] | [[File:GFP.jpg]] | ||
| + | ::::::<font size=4> [Figure 1]</font> | ||
| style="border-left:3px dotted #99F" | | | style="border-left:3px dotted #99F" | | ||
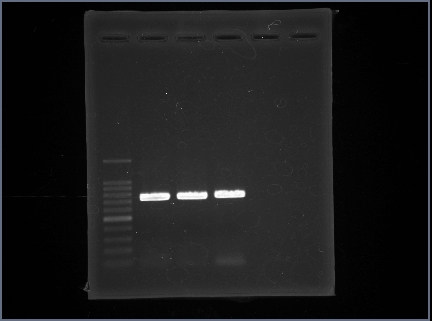
=='''<font size="5">GFP</font>'''== | =='''<font size="5">GFP</font>'''== | ||
<font size=4> | <font size=4> | ||
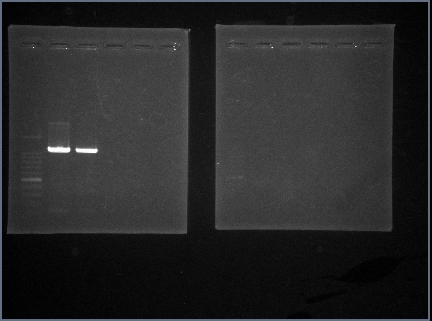
| - | We used "BBa_E0040". | + | We used "BBa_E0040". |
| - | Following figure is result of the GFP PCR. | + | Length of "BBa_E0040"is 920bp. |
| - | As a result,We were able to confirm a band about 920bp by PCR. | + | Following figure 1 is result of the GFP PCR. |
| + | As a result,We were able to confirm a band about 920bp by GFP PCR. | ||
</font> | </font> | ||
|} | |} | ||
| Line 72: | Line 74: | ||
<br> | <br> | ||
| - | == | + | {| align="left" style="width:100%; margin-right:0.2em; border-color:#99F; border-style:dotted; border-collapse:collapse; background-color:#FFF;" |
| + | | style="width:150px" | | ||
| + | [[File:HlyA.jpg]] | ||
| + | ::::::<font size=4> [Figure 2]</font> | ||
| + | | style="border-left:3px dotted #99F" | | ||
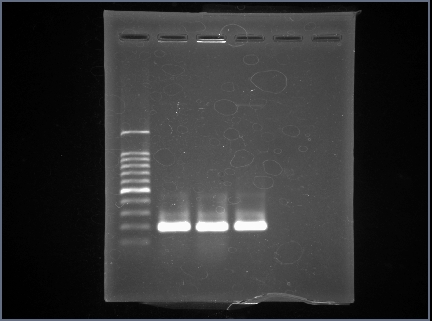
| + | =='''<font size="5">HlyA</font>'''== | ||
| + | <font size=4> | ||
| + | We used "BBa_K554002". | ||
| + | Length of "BBa_K554002" is 186bp. | ||
| + | Following figure 2 is result of the HlyA PCR. | ||
| + | As a result,We were able to confirm a band about 186bp by HlyA PCR. | ||
| + | </font> | ||
| + | |} | ||
<br> | <br> | ||
<br> | <br> | ||
| - | == | + | {| align="left" style="width:100%; margin-right:0.2em; border-color:#99F; border-style:dotted; border-collapse:collapse; background-color:#FFF;" |
| + | | style="width:150px" | | ||
| + | [[File:HlyA.jpg]] | ||
| + | ::::::<font size=4> [Figure 3]</font> | ||
| + | [[File:GFP+HlyA.jpg]] | ||
| + | ::::::<font size=4> [Figure 4]</font> | ||
| + | | style="border-left:3px dotted #99F" | | ||
| + | =='''<font size="5">GFP+HlyA and HlyA+GFP</font>'''== | ||
| + | <font size=4> | ||
| + | GFP ligated with HlyA.Length of GFP+HlyA is 920bp. | ||
| + | Following figure 3 is GFP+HlyA. | ||
| + | Following figure 4 is HlyA+GFP. | ||
| + | We submitted these parts. | ||
| + | But the function of these parts,we were not able to confirm it. | ||
| + | Because we did not have time to experiment. | ||
| + | |||
| + | </font> | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | <H1>Creating parts of STAT3</H1> | ||
| + | <font size="4"> | ||
| + | STAT3's sequence includes three recognition sequences of restriction enzyme. | ||
| + | We try to mutant this sequence,and we could succeed to mutant about one place. | ||
| + | </font> | ||
<br> | <br> | ||
<br> | <br> | ||
| - | + | ||
| + | |||
| + | <H1>Creating parts of IL-10α,IL-10β</H1> | ||
| + | <font size="4"> | ||
| + | IL-10a's sequence includes six,IL-10b's sequence includes three recognition sequences of restriction enzyme. | ||
| + | We try to mutant these sequences ,but we didn't succeed. | ||
| + | </font> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | |||
| + | |||
| + | <H1>Creating parts of HRV,IL-5Ra,IL-5Rb,AraC,Arac Promoter</H1> | ||
| + | |||
| + | {| align="left" style="width:100%; margin-right:0.2em; border-color:#99F; border-style:dotted; border-collapse:collapse; background-color:#FFF;" | ||
| + | | style="width:150px" | | ||
| + | [[File:140927 Arac AracP HRV colonyPCR.jpg]] | ||
| + | ::::::<font size=4> [Figure 1]</font> | ||
| + | [[File:140929 araC araC promoter TAvector colonyPCR.jpg]] | ||
| + | ::::::<font size=4> [Figure 2]</font> | ||
| + | | style="border-left:3px dotted #99F" | | ||
| + | |||
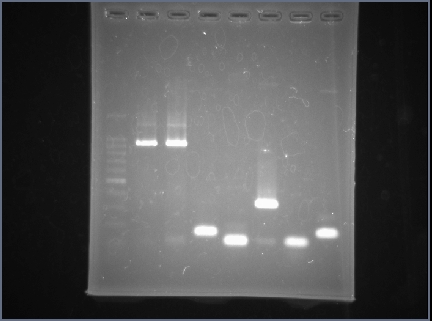
| + | =='''<font size="5"> Result of colony PCR </font>'''== | ||
| + | <font size=4> | ||
| + | Figure 1 is colony PCR of colony transformed TA vector. | ||
| + | Lane 1,2 are colony transformed TA vector inserted AraC gene. | ||
| + | Lane 3,4 are colony transformed TA vector inserted AraC promoter gene. | ||
| + | Lane 5,6,7 are colony transformed TA vector inserted HRV gene. | ||
| + | The band of lane 5 was 325bp. | ||
| + | Objective band size was 325bp. | ||
| + | We are able to confirm that we get colony which has TA vecter inserted AraC promoter gene. | ||
| + | <br> | ||
| + | <br> | ||
| + | Figure 2 is colony PCR of colony transformed TA vector inserted AraC gene. | ||
| + | The band of lane 1,2 were 1104bp. | ||
| + | Objective band size was 1149bp. | ||
| + | We are able to confirm that we get colony which has TA vector inserted AraC gene. | ||
| + | |||
| + | </font> | ||
| + | |} | ||
Latest revision as of 09:49, 17 October 2014
|
|
|
|
|
|
|
|
|
Results
GFP+HlyA(BBa_K1414000) and HlyA+GFP(BBA_K1414001)
Creating parts of STAT3
STAT3's sequence includes three recognition sequences of restriction enzyme.
We try to mutant this sequence,and we could succeed to mutant about one place.
Creating parts of IL-10α,IL-10β
IL-10a's sequence includes six,IL-10b's sequence includes three recognition sequences of restriction enzyme.
We try to mutant these sequences ,but we didn't succeed.
Creating parts of HRV,IL-5Ra,IL-5Rb,AraC,Arac Promoter
|
 "
"