Team:SUSTC-Shenzhen/Notebook/Biobricks Characterization
From 2014.igem.org
Zhangysh1995 (Talk | contribs) |
Zhangysh1995 (Talk | contribs) |
||
| Line 25: | Line 25: | ||
='''Plasmid Construction'''= | ='''Plasmid Construction'''= | ||
ALL ABBREVIATIONS USED: | ALL ABBREVIATIONS USED: | ||
| - | {| class="table" | + | {|class="table" |
! Parts name | ! Parts name | ||
! Abbreviations | ! Abbreviations | ||
| Line 200: | Line 200: | ||
|} | |} | ||
| - | ==='''DNA Purification'''=== | + | ==='''DNA Purification'''=== |
Follow instructions in kit. | Follow instructions in kit. | ||
| - | ==='''Ligation'''=== | + | ==='''Ligation'''=== |
To complete construction quickly, we use 3A assembly to achieve plasmid with resistant to chloramphenicol (A) and standard assembly with resistant to Ampicillin (B). <br> | To complete construction quickly, we use 3A assembly to achieve plasmid with resistant to chloramphenicol (A) and standard assembly with resistant to Ampicillin (B). <br> | ||
Third step ligation - plan A(3A assembly) | Third step ligation - plan A(3A assembly) | ||
| Line 257: | Line 257: | ||
! | ! | ||
! B31 | ! B31 | ||
| - | + | E10 | |
! B31 K916 | ! B31 K916 | ||
! B31 K09 | ! B31 K09 | ||
! B34 | ! B34 | ||
| - | + | E10 | |
! B34 K916 | ! B34 K916 | ||
! B34 | ! B34 | ||
| - | + | K09 | |
! B34 K11 | ! B34 K11 | ||
|- | |- | ||
| Line 282: | Line 282: | ||
| colspan="7" | 1.0 | | colspan="7" | 1.0 | ||
|- | |- | ||
| - | | T4 Ligase | + | | T4 Ligase(μL) |
| - | + | ||
| colspan="7" | 0.5 | | colspan="7" | 0.5 | ||
|- | |- | ||
| - | | T4 Ligase | + | | T4 Ligase buffer(μL) |
| - | + | ||
| colspan="7" | 1 | | colspan="7" | 1 | ||
|- | |- | ||
| Line 331: | Line 329: | ||
==='''Religation'''=== | ==='''Religation'''=== | ||
| - | {| class="table" | + | {|class="table" |
! | ! | ||
! B34 E10 | ! B34 E10 | ||
| Line 380: | Line 378: | ||
{{SUSTC-Image|wiki/images/8/82/SUSTC-Shenzhen_Characterization_10.2_electrophoresis.jpg | characterization}} | {{SUSTC-Image|wiki/images/8/82/SUSTC-Shenzhen_Characterization_10.2_electrophoresis.jpg | characterization}} | ||
| - | ==='''Enzyme | + | ==='''Enzyme Digestion I'''=== |
{| class="table" | {| class="table" | ||
! | ! | ||
| Line 565: | Line 563: | ||
|- | |- | ||
| Promoter | | Promoter | ||
| - | + | Backbone(μL) | |
| colspan="8" | 1.3 | | colspan="8" | 1.3 | ||
| 0.9 | | 0.9 | ||
Revision as of 09:10, 17 October 2014
Notebook
Biobricks Characterization
Scheme
At first, we want to characterize plasmid assembled by 3 promoters, 3 RBSs, and 4 chromoprotein (36). Because time limits, we choose 2 promoter, 2 RBSs and 4 chromoprotein (16). In carrying out experiments, we cannot easily differ new constructed plasmid with BBa_E1010 with the self-assembly one. We abandoned BBa_E1010 and do experiments on other chromoproteins.
Results
We successfully constructed 8 parts, and they all are characterized. And 6 parts were sent to Registry of Standard Biological Parts. See them | HERE.

Procedures
- Amplification of Biobricks
- Add RBS
- Add promoter
- Add terminator
Plasmid Construction
ALL ABBREVIATIONS USED:
| Parts name | Abbreviations | Parts name | Abbreviations |
|---|---|---|---|
| BBa_J23100 | J00 | BBa_E1010 | E10 |
| BBa_J23106 | J06 | BBa_K592009 | K09 |
| BBa_B0031 | B31 | BBa_K592011 | K11 |
| BBa_B0034 | B34 | BBa_K1033916 | K916 |
| BBa_B0015 | B15 | BBa_I20260 | None |
| BBa_J04450 | None |
9.29
After RBS added, all seven plasmid were cut and ligated with two promoter, J23101 and J23106 respectively.
Enzyme digestion
For plan A
| J00 | J06 | B31E10 | B31K916 | B31K09 | B34E10 | B34K916 | B34 K09 | B34K11 | |
|---|---|---|---|---|---|---|---|---|---|
| EcoRI-HF(μL) | 1 | ||||||||
| XbaI(μL) | 1 | ||||||||
| PstI | 1 | ||||||||
| EcoRV-HF | 1 | ||||||||
| NcoI | 1 | 1 | |||||||
| Linearized
Backbone(μL) | 1 | ||||||||
| DNA(μL) | 3 | 4 | 8 | 7 | 4 | 5 | 5 | 5 | 5 |
| 10X NEB Buffer 2.1(μL) | 5 | ||||||||
| ddH2O (μL) | 39 | 38 | 34 | 35 | 38 | 37 | 37 | 37 | 37 |
| Total(μL) | 40 | ||||||||
For plan B
| J00 | J06 | B31
E10 | B31
K916 | B31
K09 | B34
E10 | B34
K916 | B34 K09 | B34
K11 | |
|---|---|---|---|---|---|---|---|---|---|
| EcoRI-HF(μL) | 1 | ||||||||
| XbaI(μL) | 1 | ||||||||
| PstI | 1 | ||||||||
| NcoI | 1 | 1 | 1 | ||||||
| Linearized
Backbone(μL) | 1 | ||||||||
| DNA(μL) | 3 | 4 | 8 | 7 | 4 | 5 | 5 | 5 | 5 |
| 10X NEB
Buffer 2.1(μL) | 5 | ||||||||
| ddH2O (μL) | 39 | 38 | 34 | 35 | 38 | 37 | 37 | 37 | 37 |
| Total(μL) | 40 | ||||||||
DNA Purification
Follow instructions in kit.
Ligation
To complete construction quickly, we use 3A assembly to achieve plasmid with resistant to chloramphenicol (A) and standard assembly with resistant to Ampicillin (B).
Third step ligation - plan A(3A assembly)
| B331E10 | B31K916 | B31K09 | B34E10 | B34K916 | B34K09 | B34K11 | |
|---|---|---|---|---|---|---|---|
| DNA(50μg) | 4.0μL | 4.0μL | 2.0μL | 4.0μL | 2.0μL | 2.0μL | 4.0μL |
| 10x T4 Ligase
Buffer | 2.0μL | ||||||
| T4 Ligase | 1.0μL | ||||||
| ddH2O | 7.0μL | 7.0μL | 9.0μL | 7.0μL | 9.0μL | 9.0μL | 7.0μL |
| J23100(50μg) | 2.0μL | ||||||
| J23106(50μg) | 2.0μL | ||||||
| Backbone(50μg) | 2.0μL | ||||||
| Total | 10μL | ||||||
Third step ligation -planB(Standard Assembly)
| B31
E10 | B31 K916 | B31 K09 | B34
E10 | B34 K916 | B34
K09 | B34 K11 | |
|---|---|---|---|---|---|---|---|
| DNA(μL) | 0.7 | 0.5 | 1.2 | 0.8 | 1.2 | 1.4 | 0.7 |
| J23100(μL) | 1.2 | ||||||
| J23106(μL) | 1.0 | ||||||
| T4 Ligase(μL) | 0.5 | ||||||
| T4 Ligase buffer(μL) | 1 | ||||||
| ddH2O(μL) | 6.6 6.8 | 6.8
7.0 | 6.1
6.3 | 6.5
6.7 | 6.1
6.3 | 5.9
6.1 | 6.6
6.8 |
| Total(μL) | 10 | ||||||
Ligation: In PCR system, 16 to ligate, 65℃ to inactive, and store at 4℃.
Transformation
- Place 7 EP tubes of 100μL DH5α competent cells on ice from -80℃ to melt.
- Transfer 50μL competent cells to 7 new sterilized EP tubes from each tubes in 1.
- Add 10μL of DNA to one EP tube with competent cells respectively.
- Put all EP tubes on ice for 30mins.
- Incubate in water at 42℃ for 90 seconds, then immediately on ice for 2 minutes.
- Add 200μL SOC broth, then put in a shaking incubator for 40 minutes at 37℃ , 220rpm.
- Centrifuge at 4500rpm for 2minutes, dispose 200μL supernatant.
- Resuspend competent cells and spread plates.
Incubate at 37℃.
9.30
I. 4 plates grew single colonies
J00 B34 E10 J00 B34 K916 J06 B31 K09 J06 B34 K09
Religation
| B34 E10 | B31 K09 | B34 K09 | |
|---|---|---|---|
| DNA(50μg) | 0.8μL | ||
| J23106 | 1.0μL | 1.0μL | |
| J23100 | 1.2μL | ||
| Buffer | 1μL | 1μL | 1μL |
| Ligase | 0.5μL | 0.5μL | 0.5μL |
| ddH2O | 6.5μL | 6.3μL | 6.1μL |
| Total | 20μL | ||
===Transformation===
III. Isolate three colonies from plates of J06 B32 E10, J06 B31 K916, J06 B34 E10 and J00 B31 K09 to three tubes of 3ml LB broth respectively.
10.1
Plasmid extraction
J06 B31 K916, J00 B34 K916, J06 B34 K916, J06 B34 E10X2, J06 B31 E10 and J06 B31 K916. (Protocols following instructions in kit.) All these seven plasmid were digested with EcoRI, PstI, EcoRI-HF&PstI and go gel electrophoresis tests on 10.2. (Sample list has the same order in former text)
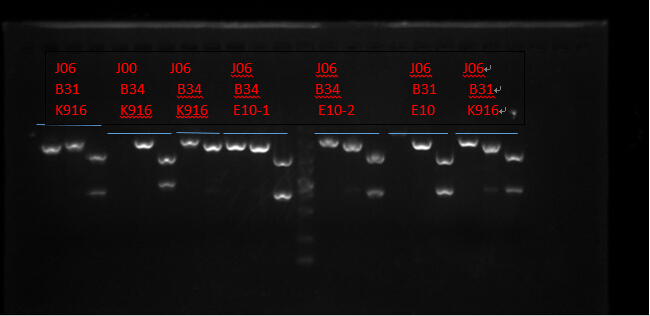
Enzyme Digestion I
| J23100 | J23106 | B31E10 | B31K916 | B31K09 | B34E10 | B34K916 | B34K09 | B34K11 | |
|---|---|---|---|---|---|---|---|---|---|
| DNA(1μg | 3.5μL | 4.1μL | 7.5μL | 6.7μL | 4.4μL | 4.5μL | 4.6μL | 4.6μL | 4.7μL |
| NEB Buffer 2.1 | 5μL | 5μL | 5.0μL | 5.0μL | 5.0μL | 5.0μL | 5.0μL | 5.0μL | 5.0μL |
| XbaI | 0.6μL | ||||||||
| SpeI | 0.6μL | 0.6μL | |||||||
| PstI | 0.6μL | ||||||||
| ddH2O | 40.3μL | 39.7μL | 36.3μL | 37.1μL | 39.4μL | 39.3μL | 39.2μL | 39.2μL | 39.1μL |
| Total | 50μL | ||||||||
Digest Overnight
Enzyme Digestion II
| J06 B31 K916 | J00 B34 K916 | J06 B34 K916 | J06 B34 E10-1 | J06 B34 E10-2 | J06 B31 E10 | J06 B31 K916 | |
|---|---|---|---|---|---|---|---|
| DNA(μL) | 2 | ||||||
| Buffer(μL) | 1 | ||||||
| EcoRI-HF(μL) | 0.3 | ||||||
| PstI(μL) | 0.3 | ||||||
| ddH2O(μL) | 6.4 | ||||||
| Total(μL) | 10 | ||||||
Digest overnight
Enzyme Digestion III
| pSB1C3 RFC | B31K09 | B31 K916 | B31 K09 | B34 E10 | B34 | B34 K11 | B34 | K11 | B31 | J00 | J06 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| XbaI(μL) | 2 | |||||||||||
| PstI(μL) | 2 | |||||||||||
| SpeI(μL) | 2 | |||||||||||
| DNA(μL) | 12 | 15 | 13 | 9 | 9 | 10 | 10 | 10 | 10 | 11 | 7 | 9 |
| 10X NEB Buffer 2.1(μL) | 10 | |||||||||||
| ddH2O(μL) | 74 | 71 | 73 | 77 | 77 | 76 | 76 | 76 | 76 | 75 | 79 | 77 |
| Total | 100 | |||||||||||
Digest overnight.
10.2
Gel electrophoresis
54μL reaction,9μL loading dye
Gel extraction (10.1 II)
Ligation (T)
| J00 B31 E10 | J00 B31 K09 | J00 B34 E10 | J00 B34 K09 | J00 B34 K11 | J06 B31 E10 | J06 B31 K09 | J06 B34 E10 | B31 K11 | J00 | J06 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ligase(μL) | 0.5 | ||||||||||
| Buffer(μL) | 1 | ||||||||||
| Promoter
Backbone(μL) | 1.3 | 0.9 | 1.3 | ||||||||
| DNA(μL) | 1.2 | 1.5 | 1.6 | 1.6 | 1.6 | 1.2 | 1.5 | 1.6 | 1.4 | ||
| ddH2O(μL) | 6 | 5.7 | 5.6 | 5.6 | 5.6 | 6 | 5.7 | 5.6 | 6.2 | 7.2 | 7.2 |
| Total(μL) | 10 | ||||||||||
Isolate three colonies from plates of J00 B31 K916, J06 B31 K916, J06 B34 K916, J06 B34 K09, J06 B34 K11 to tubes with 3mL LB broth respectively.
References
- [http://www.tiangen.com/en/?productShow/t1/4/id/32.html |TIANprep Mini Plasmid Kit]
- [http://www.tiangen.com/en/?productShow/t1/4/id/41.html |TIANprep Midi Purification Kit]
- |NEB Biobricks® Assembly Kit
 "
"