Team:ETH Zurich/lab/biobrick/used1
From 2014.igem.org
(→Experimental Set-Up) |
(→Experimental Set-Up) |
||
| Line 15: | Line 15: | ||
=== Experimental Set-Up === | === Experimental Set-Up === | ||
| - | The above described ''E. coli'' TOP10 strain was grown overnight in Lysogeny Broth (LB) containing kanamycin (50 μg/mL) and ampicillin (200 μg/mL) to an OD<sub>600</sub> of about 1.5. As a reference, a preculture of the same strain lacking the sfGFP gene was included. The cultures were then diluted 1:40 in fresh LB containing the appropriate antibiotics and measured in microtiter plate format on 96-well plates (200 μL culture volume) with a Tecan infinite M200 PRO plate reader (excitation | + | The above described ''E. coli'' TOP10 strain was grown overnight in Lysogeny Broth (LB) containing kanamycin (50 μg/mL) and ampicillin (200 μg/mL) to an OD<sub>600</sub> of about 1.5. As a reference, a preculture of the same strain lacking the sfGFP gene was included. The cultures were then diluted 1:40 in fresh LB containing the appropriate antibiotics and measured in microtiter plate format on 96-well plates (200 μL culture volume) for 10 h with a Tecan infinite M200 PRO plate reader (optical density at 600 nm; fluorescence with an excitation wavelength of 488 nm and an emission wavelength of 530 nm). |
=== Modeling leakiness === | === Modeling leakiness === | ||
Revision as of 19:55, 16 October 2014
|
<partinfo>BBa_R0062 AddReview 4</partinfo> ETH Zurich 2014 |
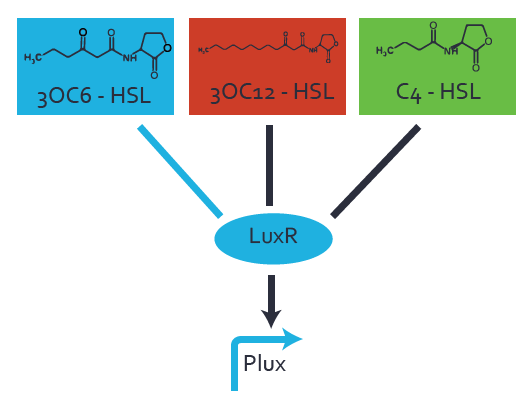
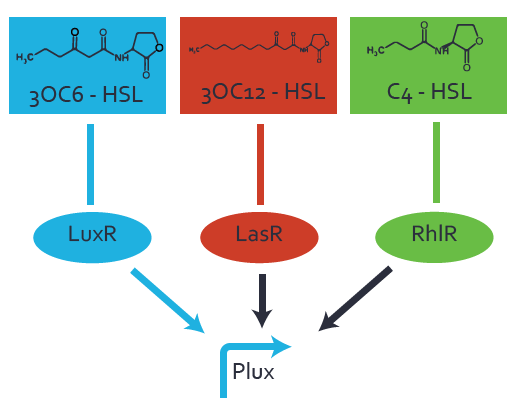
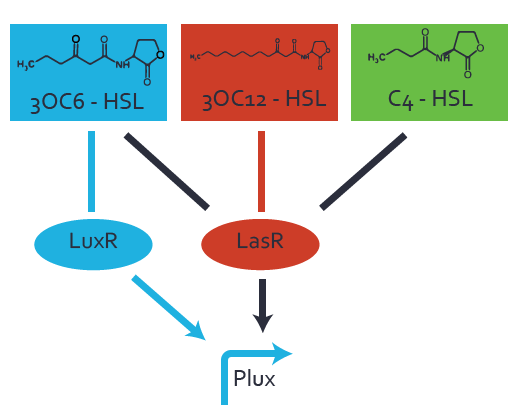
Characterization of the promoter's basal leakiness to LuxR in the absence of 3OC6-HSLThe amount of regulator [http://parts.igem.org/Part:BBa_C0062:Experience LuxR (BBa_C0062)] in the system was shown to influence the pLuxR promoter's basal expression or leakiness. By using the three different constitutive promoters [http://parts.igem.org/Part:BBa_J23100:Experience BBa_J23100], [http://parts.igem.org/Part:BBa_J23109:Experience BBa_J23109], and [http://parts.igem.org/Part:BBa_J23111:Experience BBa_J23111] for the production of LuxR we have measured this effect in terms of fluorescence. Background informationWe used an E. coli TOP10 strain transformed with two medium copy plasmids (about 15 to 20 copies per plasmid and cell). The first plasmid contained the commonly used p15A origin of replication, a kanamycin resistance gene, and promoter [http://parts.igem.org/Part:BBa_R0062 pLuxR (BBa_R0062)] followed by [http://parts.igem.org/Part:BBa_B0034 RBS (BBa_B0034)] and superfolder green fluorescent protein (sfGFP). In general, for spacer and terminator sequences the parts [http://parts.igem.org/Part:BBa_B0040 BBa_B0040] and [http://parts.igem.org/Part:BBa_B0015 BBa_B0015] were used, respectively. The second plasmid contained the pBR322 origin (pMB1), which yields a stable two-plasmid system together with p15A, an ampicillin resistance gene, one of three promoters chosen from the [http://parts.igem.org/Promoters/Catalog/Anderson Anderson promoter collection] followed by [http://parts.igem.org/Part:BBa_C0062 luxR (BBa_C0062)]. The detailed regulator construct design and full sequences (piG0041, piG0046, piG0047) are available here. Experimental Set-UpThe above described E. coli TOP10 strain was grown overnight in Lysogeny Broth (LB) containing kanamycin (50 μg/mL) and ampicillin (200 μg/mL) to an OD600 of about 1.5. As a reference, a preculture of the same strain lacking the sfGFP gene was included. The cultures were then diluted 1:40 in fresh LB containing the appropriate antibiotics and measured in microtiter plate format on 96-well plates (200 μL culture volume) for 10 h with a Tecan infinite M200 PRO plate reader (optical density at 600 nm; fluorescence with an excitation wavelength of 488 nm and an emission wavelength of 530 nm). Modeling leakinessResultsCharacterization of the promoter's sensitivity to 3OC6-HSL depending on LuxR concentrationBackground informationSystems consideredModeling promoter's sensitivityResultsCharacterization of two-level crosstalk on the promoterBackground informationSystem consideredModeling crosstalkFirst-order crosstalkFirst Level crosstalk: LuxR binds to different HSL and activates the promoterSecond Level crosstalk: other regulatory proteins, like LasR, bind to their natural HSL substrate and activates the promoterSecond order crosstalk: Combination of both cross-talk levelsOther regulatory proteins, like LasR, bind to different HSL and activates the promoter Results |
 "
"