Team:Heidelberg/pages/PCR 2.0
From 2014.igem.org
(→Material and methods) |
(→Material and methods) |
||
| Line 35: | Line 35: | ||
=Material and methods= | =Material and methods= | ||
| - | |||
| - | |||
{{:Team:Heidelberg/templates/image-quarter| | {{:Team:Heidelberg/templates/image-quarter| | ||
| Line 43: | Line 41: | ||
descr=Placeholder| | descr=Placeholder| | ||
file=Purification_procedure.png}} | file=Purification_procedure.png}} | ||
| - | |||
| - | |||
{{:Team:Heidelberg/templates/image-half| | {{:Team:Heidelberg/templates/image-half| | ||
| Line 51: | Line 47: | ||
descr=Placeholder| | descr=Placeholder| | ||
file=MethylationAssay_overview.png}} | file=MethylationAssay_overview.png}} | ||
| - | |||
| - | |||
{{:Team:Heidelberg/templates/image-half| | {{:Team:Heidelberg/templates/image-half| | ||
| Line 59: | Line 53: | ||
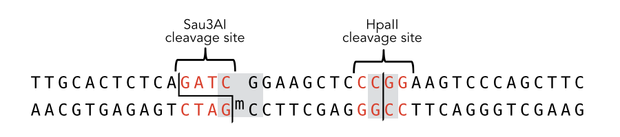
descr=Sequence of the 40-mer oligo. CpG dinucleotides are shaded in gray, Sau3AI and HpaII recognition sites are red. The first CpG is methylated on the reverse strand.| | descr=Sequence of the 40-mer oligo. CpG dinucleotides are shaded in gray, Sau3AI and HpaII recognition sites are red. The first CpG is methylated on the reverse strand.| | ||
file=DNMT1_substrate.png}} | file=DNMT1_substrate.png}} | ||
| - | |||
| - | |||
=Results= | =Results= | ||
=References= | =References= | ||
Revision as of 21:43, 12 October 2014
Contents |
Introduction
The invention of the polymerase chain reaction in 1983 by Kary Mulliy revolutionised the modern world by enabling amplification of DNA in an exponential manner. Further improvements including the use of thermo-stable DNA-polymerases made the method even more efficient, allowing its widespread use in nearly every field of modern diagnostics and research. However, this method is limited to the base pair sequence leaving large portion of information DNA methylation pattern which gives essential information about the transcriptional state of a given gene. The growing interest in such modifications that influence and define DNA functions without changing the actual sequence is in the focus of epigenetic research. One of the greatest challenge is to obtain enough source material for analysis. Currently there is no method to produce methylated DNA in a substantial amount for real life studies.
Our work presents an easier and more efficient way to produce methylated DNA in an exponential manner. We call it PCR 2.0, a PCR reaction that is not only capable of amplifying the DNA base pair sequence, but also to maintain epigenetic information on every DNA fragment. The pivotal element of this PCR reaction is the use of DNA methyltransferase, which needs to be resistant against temperatures above 60°C. Although DNA methyltransferases can be found in thermophile organisms, (5) until now no suitable protein was found or synthesised to withstand PCR conditions. Here we have applied the method of circularisation from our intein toolbox and the linker design software to create a thermo-stable DNM methyltransferase to advance the vision of a PCR 2.0.
Background
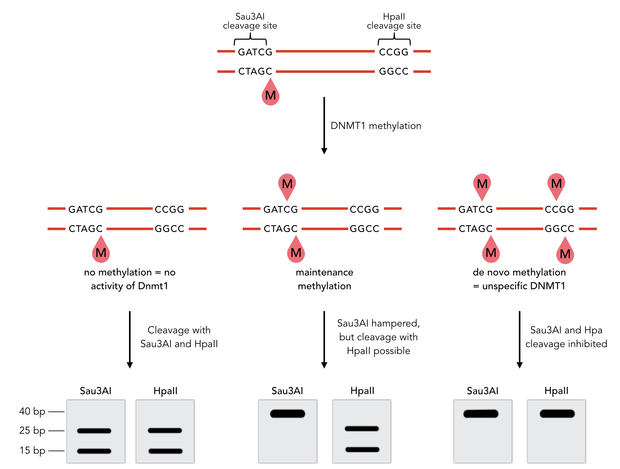
In mammals, DNA methylation plays an active role in gene regulating and cell memory. It mainly occurs at CpG dinucleotides in promoter regions or in repetitive sequences of the DNA where a methyl group (CH3) is covalently linked to the 5’ end cytosine on both complementary DNA strands. Depending on the numbers and positions of the methyl groups, gene expression can be enhanced or silenced, giving each cell an individual protein expression pattern and subsequent function. A family of enzymes called DNA methyltransferases (DNMTs) are responsible for the establishment and maintenance of cell-type specific DNA methylation patterns. The enzymes DNMT3a and DNMT3b have a role in de novo methylation of DNA during development, whereas DNMT1 preserves this methylation pattern in every cell cycle by transferring the methylation pattern from the parental DNA strand to the new daughter strand. (1)
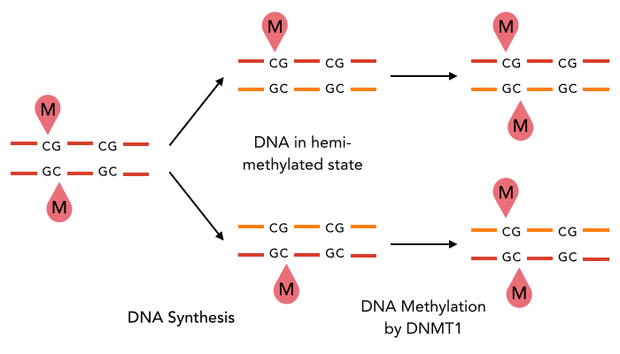
After semiconservative replication, only one DNA strand contains the epigenetic information. DNMT1 recognises hemi-methylated DNA and transfers a methyl group from the donor molecule S-adenosyl-L-methionine (SAM) to the C5 position of the cytosine residue on the complement DNA strand, creating a 5-methylcytosine.
DNA methylation is a precondition for cell differentiation and a deviation in the pattern may lead to malfunction of the cell, resulting in cancer growth, mental illnesses and other diseases (6). Large efforts were made to decipher human DNA methylation patterns. Although now it can be detected (reviewed in (7)), direct amplification of methylated DNA fragments, e.g. for functional analysis in vitro, is impossible up to now. Our project aims facilitate epigenetic research by developing a more stable DNMT1 enabling amplification of methylated DNA.
Circularisation of Dnmt1
Dnmt1 is of particular interest for a methylation maintenance PCR, as it allows exact duplication of the original methylation pattern to every new DNA copy without introducing de novo methylation.
PCR uses three different temperatures for denaturing, annealing and elongation of the DNA sequence, ranging from 55°C to 98°C, requiring extremely heat enzymes.
Circularisation is one way to increase heat stability of a protein, because it ankers the loose terminal ends to the rest of the protein. denaturation typically starts at the loose terminal ends of a protein. This decreases the high mobility of the regions and therefore making them less affected for denaturing. Further information can be found at the circularisation toolbox element.
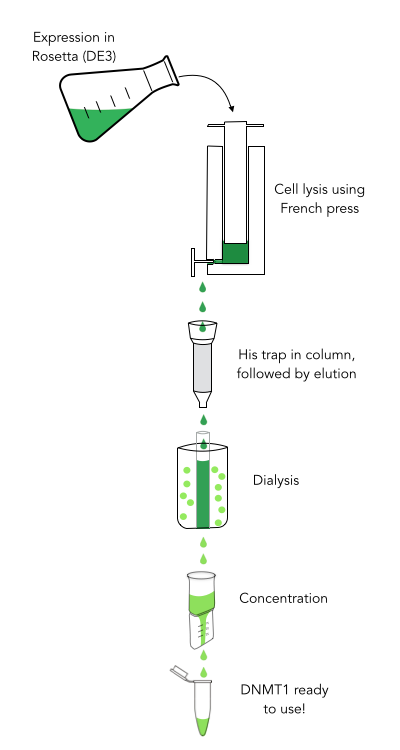
For our experiments we used a truncated version of DNA methyltransferase (cytosine-5) 1 of Mus musculus (713-1602 amino acids) kindly provided by Dr. Pavel Bashtrykov. In contrast to the full-length Dnmt1 that can only be expressed in a baculovirus-insect cell system, this truncated Dnmt1 can also be expressed in E.coli. Moreover, the N and C terminal ends of Dnmt1 (731-1602) are closer together than in the full-lengh Dnmt1. This likely facilitates circularization and might cause less deformation and a lower impact on overall activity. Dnmt1(731-1602) contains the C-terminal catalytic domain and the aligning bromo-adjacent homology (BAH) domains, which prevents the binding of unmethylated DNA in the catalytic core. The enzyme is missing the CXXC domain, which has a high affinity to CpG hemi-methylated dinucleotides, but was shown to be dispensable for protein function (2). The truncated version has almost the same methylation activity and specificity as the wild type, as shown by our own experiments and by Bashtrykov and colleagues (2).
However, even the truncated Dnmt1 is a very large protein of 100kDa and consists of 872 amino. A protein this size has never been circularised before. According to the crystal structure, the distance between the termini of the truncated Dnmt1 is 48 Å.
We suspected circularization by direct fusion of both termini might cause a strong deformation of the Dnmt1 structure leading to a loss of function. Therefore we used a linker amino acid sequence, adapted to the structure of Dnmt1: it needed to be long enough to bridge the gap of 48 Å and bypassing the catalytic core located prominently in the middle of the 3 dimensional enzyme structure.
For circularisation we used NpuDnaE Intein, very efficient inteins for splicing the terminal ends together, as we already tested in circularisation of GFP, Lysosyme and Xylanse.
We tried two different linkers: one rather rigid linker, calculated by a previous version of our linker software subsequently optimised through experience, and one flexible linker.
Material and methods
 "
"