From 2014.igem.org
(Difference between revisions)
|
|
| Line 304: |
Line 304: |
| | | | |
| | <p> | | <p> |
| - | :1)We put 1μℓeach plasmid into 3 microtubes.(used plasmid are pCold vectorⅠ(4407bp),pCold vectorⅡ(4392bp) and pSBIC3) | + | :1) |
| | </p> | | </p> |
| | | | |
| | <p> | | <p> |
| - | :2)Put 1ul PstⅠand 1ul EcoRⅠinto each microtube. | + | :2) |
| | </p> | | </p> |
| | | | |
| | <p> | | <p> |
| - | :3)Put 1ul ×10 H buffer into each microtube. | + | :3) |
| | </p> | | </p> |
| | | | |
| | <p> | | <p> |
| - | :4)Up to 10ul with D<sub>2</sub>W. | + | :4) |
| | </p> | | </p> |
| | | | |
| | <p> | | <p> |
| - | :5)Did heat block(37℃) about 2hour. | + | :5) |
| | </p> | | </p> |
| | | | |
| | <p> | | <p> |
| - | :6)Prepared 3 new microtubes.and added 10ul GFP+HlyA into them. | + | :6) |
| | </p> | | </p> |
| | | | |
| | <p> | | <p> |
| - | :7)added D<sub>2</sub>W until it became 10ul | + | :7) |
| | </p> | | </p> |
| | | | |
| | </font> | | </font> |
Revision as of 08:06, 17 September 2014

|
|
|
|
|

Protocol
|
|
|
|
|
|
|
|
Protocol
1:miniprep
- ・We took plasmid out of Escherichia coli which have a gene of IL-10 α and IL-10 β and STAT3 in miniprep to use it by a following experiment (the Escherichia coli which I really used in an experiment of 2013).
DNA of HlyA and the GFP are IGEM 2014 kit plate1 21G and IGEM 2014 kit plate 13L, so we didn't miniplep
- 1) We cultured bacterial strain with the LB medium which I added ampicillin to so that density becomes 100ug/ml overnight.(We made a nutrient medium of around 5 ml in 50 ml falcons)(Against 5 ml of nutrient mediums, Amp used 5ul)
- 2) Aliquot 1ml culture into a 1.5 ml microcentrifuge tube,and Made it spin at 10000rpm (4℃) for 1 min to harvest the bacteria.
- 3) Removed supernatant and performed 2)operation again, Removed supernatant .
- 4) Resuspended bacterial pellet by complete vortexing in 100ul SolutionⅠ{D-glucose:9g(50mM),1M Tris-HCl(pH 8.0):25ml(25mM),0.5M EDTA:20ml(10mM),H2O:955ml /1L}.
- 5) Inverted bacterial pellet by complete fall mixtureing in 200ul SolutionⅡ{NaOH:8g,SDS:10g[1%(w/v)],H2O:960ml /1L},and confirmed that it became transparent.
- 6) Cooled for three minutes in ice.
- 7) Inverted bacterial pellet by complete fall mixtureing in 150ul SolutionⅢ{CH3COOH:294.5g(3M),CH3COOH:120ml(2M),H2O:diluting in measuring cylinder to 1L total},and confirmed that it became Cloudiness.
- 8) Cooled for 3 minutes in ice.
- 9) Harvested the DNA by spinning at 10000rpm (4℃) for 10 min.
- 10) Gathered only supernatant and moved it in a new microcentrifuge tube.
- 11) Added 0.8ul Rnase(10mg/ml) and incubate the solution(37℃,1min)
- 12) Added 200ul phenol:chloroform(1:1) and inverted
- 13) Harvested by spinning at 10000rpm (4℃) for 5 min.
- 14) Removed only supernatant and moved it in a new microcentrifuge tube, after that tapped in 200ul chloroform.
- 15) Harvested by spinning at 10000rpm (4℃) for 1 min.
- 16) Moved its supernatant to a new microcentrifuge tube and add 15ul 3M CH3COONa.(Don't gather underlayer)(The ratio of the 3M sodium acetate and supernatant is made to be 10:1)
- 17) Added 400ul 100%CH3CH2OH and made it stirred well.
- 18) Harvested by spinning at 10000rpm (4℃) for 20 min.
- 19) Removed only supernatant and added 400ul 70%CH3CH2OH(pour a liquid from the other side for white thing not to drain a white.[white thing is plasmid])
- 20) Harvested by spinning at 10000rpm (4℃) for 20 min.
- 21) Removed only supernatant and opened the cover of the tube for 10min to dry CH3CH2OH.
- 22) Added 50ul TE to dissolve DNA
- 23) We stored low temperature
2:PCR
- ・We performed PCR to confirm whether DNA which we need.
- 1)We diluted the primer.(D2W:primer=4:1)
- 2)Made PCR preparation liquid {buffer×10:2.5ul,dNTP:2ul,PrimerF:1ul,PrimerR:1ul,Taq Polumerase:0.1ul,D2W:17.39ul /1 microcentrifuge tube(0.2ml)}
- 3)Added sample(there are Plasmid made with miniprep)to the microcentrifuge tube and do PCR(The PCR conditions are as follows).
- Figure①:PCR condition of STAT3 ,IL-10a and IL-10b

- [②→③→④]:repeated 35~40 cycles
- Figure②:PCR condition of GFP and HlyA
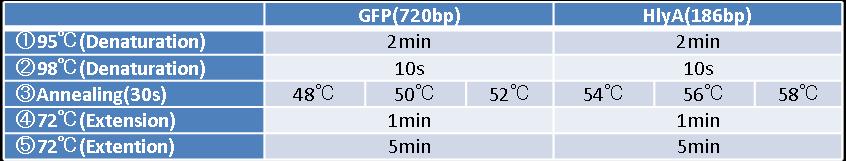
- [②→③→④]:repeated 40 cycles
- 4)We did Electrophoresis to confirm Objective band.
3:Restriction enzyme processing
- ・We did restriction enzyme processing to plasmid to incorporate inserts in a plasmid vector.
- Figure③:Used Restriction enzyme
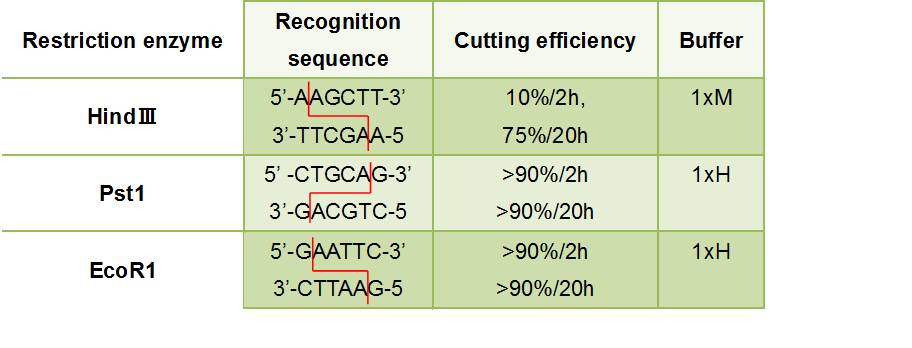
- HlyA・・・We removed the stop codon
- F chain ・・・This has EcoR1 recognition sequence.We added CG and A to this to prevent flame out.
- ・5'-CGGAATTCATTAGCCTATGGAAGTCAGGG-3'
- ((Tm before adding a restriction enzyme site =60℃
- ((GC before adding a restriction enzyme site =50.0%
- R chain ・・・This has HindⅢ recognition sequence.We added CCC to this.
- ・5'-CCCAAGCTTTGCTGATGTGGTCAGGGTTA-3'
- ((Tm before adding a restriction enzyme site =60℃
- ((GC before adding a restriction enzyme site =50.0%
- GFP
- F chain ・・・This has HindⅢ recognition sequence.We added CCC and A to this to prevent flame out.
- ・5'-CCCAAGCTTAATGCGTAAAGGAGAAGAACT-3'
- ((Tm before adding a restriction enzyme site =56℃
- ((GC before adding a restriction enzyme site =40.0%
- R chain ・・・This has Pst1 recognition sequence.We added AA to this.
- ・5'-AACTGCAGTTATTATTTGTATAGTTCATCC-3'
- ((Tm before adding a restriction enzyme site =54℃
- ((GC before adding a restriction enzyme site =22.7%
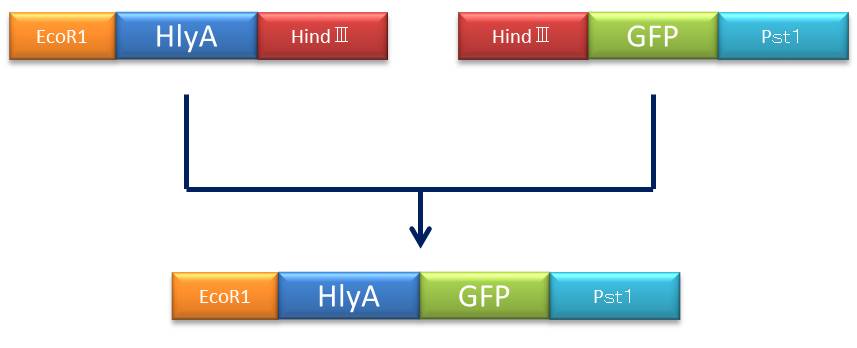
- Figure④:Rough plan
- 1)We set a heat block(37℃).
- 2)prepare one microtube.
- 3)added 1ul restriction enzyme buffer(×10) to microtube
- 4)added 1ul DNA sample liquid to the microtube
- 5)added 1ul each restriction enzyme to the microtube
- 6)added D2W until it became 10ul
- 7)set microtube to a heat block (37℃)
- 8)left the microtue more than one hour to push forward a reaction
4:Ligation
- ・We did restriction enzyme processing to plasmid to incorporate inserts in a plasmid vector.
5:Transformation
- ・We did restriction enzyme processing to plasmid to incorporate GFP+HlyA in a plasmid vector.And we incorporated GFP+HlyA in plasmid by ligation processing
- 1)
- 2)
- 3)
- 4)
- 5)
- 6)
- 7)
|
 "
"