Team:Oxford/biosensor deterministic equations
From 2014.igem.org
(Difference between revisions)
Olivervince (Talk | contribs) |
Olivervince (Talk | contribs) |
||
| Line 264: | Line 264: | ||
| - | <div style="background-color:#D9D9D9; opacity:0.7; z-index:5; margin-right:auto;margin-left:auto; Height:75px; width: | + | <div style="background-color:#D9D9D9; opacity:0.7; z-index:5; margin-right:auto;margin-left:auto; Height:75px; width:84%;min-width:300px;font-size:65px;font-family:Helvetica;padding-top:5px; font-weight: 450;"> |
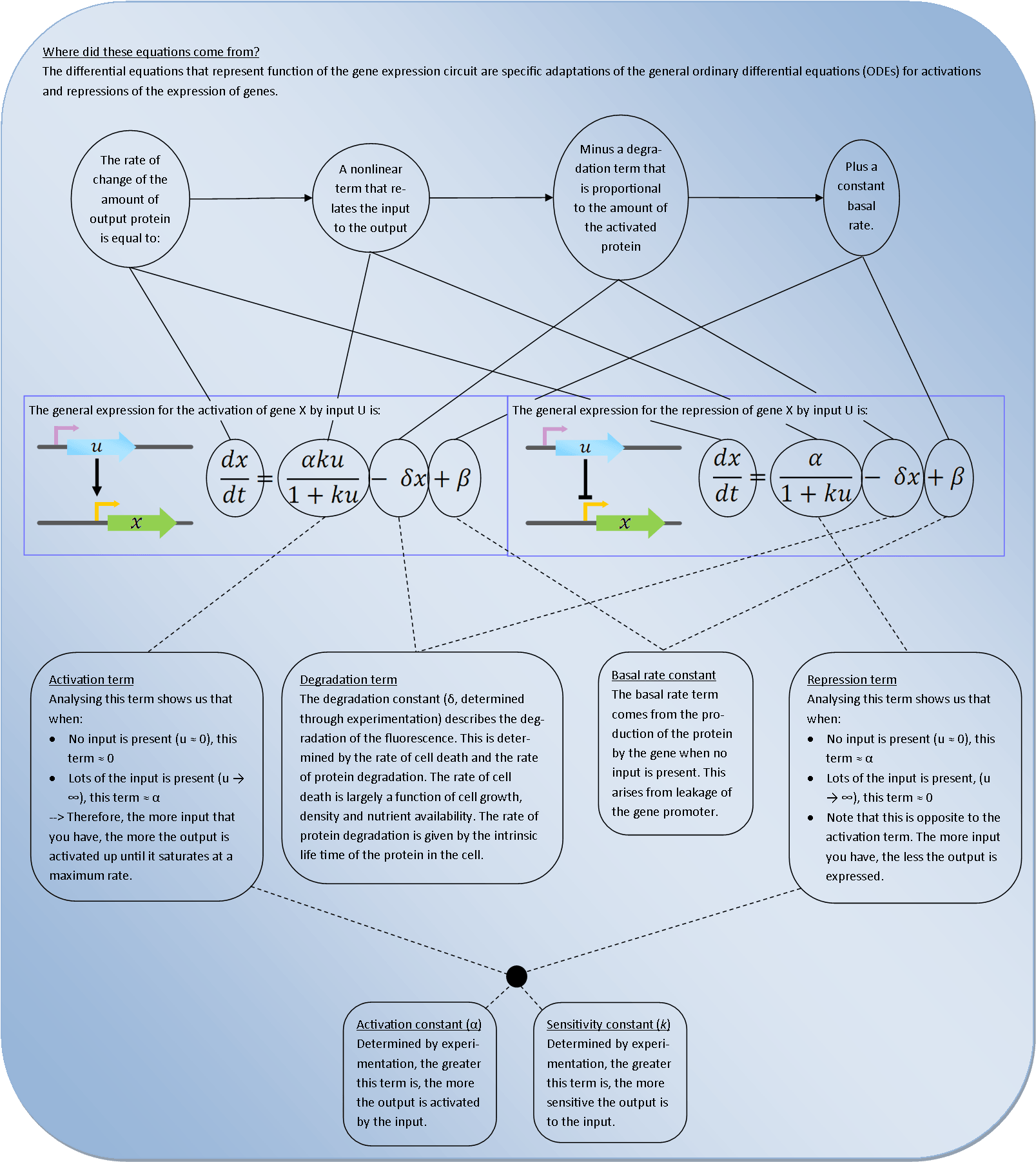
<div style="background-color:white; opacity:0.7; Height:75px; width:100%;margin-top:5px:margin-bottom:5px;min-width:300px;font-size:65px;font-family:Helvetica;padding-top:5px; color:#596C8A; font-weight: 450;"><br>Activator/Repressor equations</div> | <div style="background-color:white; opacity:0.7; Height:75px; width:100%;margin-top:5px:margin-bottom:5px;min-width:300px;font-size:65px;font-family:Helvetica;padding-top:5px; color:#596C8A; font-weight: 450;"><br>Activator/Repressor equations</div> | ||
</div> | </div> | ||
Revision as of 23:59, 16 September 2014
 "
"