Team:Tianjin/Project
From 2014.igem.org
(Difference between revisions)
| Line 353: | Line 353: | ||
<p> </p> | <p> </p> | ||
<p> </p></td> | <p> </p></td> | ||
| - | <td width="43%" bgcolor="#3A2108"><p><span class="STYLE43"><span class="STYLE24"><a name="top" id="top"></a><span class="STYLE77">T</span></span><span class="STYLE76">rans<em>f</em> ibre</span></span><br /> | + | <td width="43%" bgcolor="#3A2108"><p> </p> |
| - | + | <p> </p> | |
| - | + | <p> </p> | |
| + | <p><span class="STYLE43"><span class="STYLE24"><a name="top" id="top"></a><span class="STYLE77">T</span></span><span class="STYLE76">rans<em>f</em> ibre</span></span><br /> | ||
| + | </p> | ||
| + | <p> </p> | ||
| + | <p><span class="STYLE40">Welcome to Team Tianjin! </span><br /> | ||
| + | </p></td> | ||
<td width="38%" bgcolor="#3A2108"><img src="https://static.igem.org/mediawiki/2014/e/eb/Tianjin-head3.gif" width="456" height="226" align="right" /></td> | <td width="38%" bgcolor="#3A2108"><img src="https://static.igem.org/mediawiki/2014/e/eb/Tianjin-head3.gif" width="456" height="226" align="right" /></td> | ||
</tr> | </tr> | ||
| Line 422: | Line 427: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td bgcolor="#FFED97" class="STYLE33"><p> | + | <td bgcolor="#FFED97" class="STYLE33"><p>As to lower organisms, there seems an indestructible barrier between biological signals and electrical signals. There have been bits of research results on the transformation from electrical signals to biological signals, for example, the phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor in Ciona intestinalis. However, the opposite direction of transformation is almost a blank space, especially in microbes. Thus, we want to build a bridge between them. With this, microbes can accomplish detection with electronic output signals, and even ‘operate’ machines!</p> |
| - | <p> | + | <p>The bridge is the transformed curli fiber linked to nanogold. When inducer is added in the system, ‘transfibers’ take shape and nanogolds are linked together as wires, so the circuit is ‘on’. In terms of detection, it can be characterized by electric current change. Meanwhile, other machines can be switched on by means of this.</p> |
| - | <p> | + | <p>To learn more, follow us and enjoy the travel of ‘TRANS’!</p></td> |
</tr> | </tr> | ||
</table></td> | </table></td> | ||
| Line 433: | Line 438: | ||
<tr> | <tr> | ||
<td bgcolor="#FFED97"><p class="STYLE33"><strong>Curli fiber</strong></p> | <td bgcolor="#FFED97"><p class="STYLE33"><strong>Curli fiber</strong></p> | ||
| - | <p class="STYLE33"><strong> | + | <p class="STYLE33"><strong>1.What is curli fiber?</strong></p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">Bacteria are able to integrate and survive in a remarkably diverse collection of environments.</p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">In recent years, bacterial communities have been better appreciated as an integral part of most microbial lifestyles. These communities, or biofilms, are prominent during infections and are generally characterized by an extracellular matrix that can help sculpt three-dimensional structures, which promote the survival of its inhabitants in the face of environmental stresses. Enteric bacteria such as Escherichia coli and Salmonella spp. express proteinaceous extracellular fibers called “<strong>curli</strong>” that are involved in surface and cell-cell contacts that promote community behavior and host colonization.</p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">Curli are the major proteinaceous component of a complex extra-cellular matrix. Curli fibers are involved in adhesion to surfaces, cell aggregation and biofilm formation. Curli also mediate host cell adhesion and invasion, and they are potent inducers of the host inflammatory response.</p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">Structurally and biochemically, curli belong to a growing class of fibers known as amyloids. Amyloid fiber formation is responsible for several human diseases including Alzheimer's, Huntington's, and prion diseases. Curli share all of the biophysical properties of amyloids: ordered β-sheet-rich fibers resistant to proteases and other harsh denaturants with a capacity to bind the dyes CR and ThT. In contrast to disease-associated amyloids, curli assembly is not the result of protein misfolding. Instead, curli are the product of a dedicated biogenesis pathway providing a paradigm for understanding controlled amyloidogenesis. </p> |
| - | <p class="STYLE33"><strong> | + | <p class="STYLE33"><strong>2.Why do we choose curli?</strong> </p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">Curli assembly is guided by the products of seven curlispecific genes (csg) encoded on two divergently transcribed operons, <em>csgDEFG</em> and <em>csgBAC</em>. CsgD is the master regulator of curli biogenesis and is required for transcription of the csgBAC operon.</p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">Our model system is based on the self-assembly of the secreted major curli subunit CsgA. Secreted CsgA monomers are templated on CsgB, which is anchored to the cell surface, to form curli fibre; moreover, CsgA secreted from one cell can interact with CsgB on other cells. Situated at the cell surface, CsgB nucleates soluble, unstructured CsgA into a highly ordered amyloid fiber. </p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">To transport the major submit CsgA and the minor submit CsgB into the extracellular milieu, the lipoprotein CsgG oligomerizes into a pore-like structure in the outer membrane.<br /> |
In addition, the secretion and localization of curli fiber subunits through CsgG are mediated by three additional proteins: CsgC, CsgE and CsgF. CsgC was predicted to have oxidoreductase activity, regulating CsgG outer membrane assembly and pore activity. CsgE could be considered a CsgA-specific chaperone. Besides, CsgF associates with the outer membrane and is required for cell association of the minor curli fiber subunit CsgB.</p> | In addition, the secretion and localization of curli fiber subunits through CsgG are mediated by three additional proteins: CsgC, CsgE and CsgF. CsgC was predicted to have oxidoreductase activity, regulating CsgG outer membrane assembly and pore activity. CsgE could be considered a CsgA-specific chaperone. Besides, CsgF associates with the outer membrane and is required for cell association of the minor curli fiber subunit CsgB.</p> | ||
| - | <p class="STYLE33"> | + | <p class="STYLE33">As explained above, curli have two remarkable advantages. One is the self-assembly of CsgA monomers into mature amyloids----curli. Apart from it, using synthetic riboregulators, we can implement inducible transcriptional and translational control over the expression of CsgA subunits engineered to display various peptide tags, which can interface with inorganic materials, such as gold nanoparticles, to form a conductive structure. Utilizing these advantages, we can design a transducer which can convert the change of gene expression level that stimulated by inducer directly into the electric signal via inductive synthesis(or destroy)of nanowire between the electrodes.</p> |
<p class="STYLE33"><strong>Gold Nanoparticles (AuNPs)</strong> </p> | <p class="STYLE33"><strong>Gold Nanoparticles (AuNPs)</strong> </p> | ||
| - | <p class="STYLE33"> | + | <p class="STYLE33">AuNPs are the most stable metal nanoparticles, and they present fascinating aspects such as their assembly of multiple types involving materials science, the behavior of the individual particles, size-related electronic, magnetic and optical properties (quantum size effect), and their applications to catalysis and biology.</p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">Since we can interface curli fiber with inorganic materials to produce amyloid-based wires that are either externally controllable or undergo autonomous patterning, a material with completely bio-compatibility, conductivity and stability is required. AuNPs can be linked to some peptides, providing fascinating propoties. For instance, the assembly of relatively short polypeptides on curli can provide protein-like complex structures and conductivity in conjugates with AuNPs.</p> |
<p class="STYLE33"><strong>Nanowire-based devices</strong></p> | <p class="STYLE33"><strong>Nanowire-based devices</strong></p> | ||
| - | <p class="STYLE33"> | + | <p class="STYLE33">Devices based on nanowires have emerged as one of the most powerful and general platforms for ultrasensitive, direct electrical detection of biological and chemical species and for building functional interfaces to biological systems. With collective efforts and creation of researchers all around the world, there comes many kinds of nanosensors for various uses such as ultrasensitive detection of proteins and individual virus particles as well as recording, stimulation, and inhibition of neuronal signals in nanowire–neuron hybrid structures.</p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">The advantages of nanowires lie in many ways. Firstly, nanowires are enabled reproducible synthesis of nanowires of homogeneous composition and diameter with controllable electronic and optical properties. Moreover, branched nanowire structures with unique functions have been built in at the stage of synthesis. Significantly, In general, the similarity in size of nanowires and natural nanostructures in biological systems makes nanowires an obvious choice for creating highly sensitive tools that can probe biological systems. The great characteristic of direct and label-free readout (i.e., without the use of bound dyes and fluorescent probes) is particularly attractive for many applications in medicine and life sciences.</p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">An important point about this detection process, which is quite distinct from common optically based assays, is that it occurs in real time, and the binding process can be viewed as it happens on a computer logging the conductance of one or more devices.</p> |
<p class="STYLE33"><strong>What is MMP-7?</strong></p> | <p class="STYLE33"><strong>What is MMP-7?</strong></p> | ||
| - | <p class="STYLE33"> | + | <p class="STYLE33">Matrilysin, also known as matrix metalloproteinase-7 (MMP-7), pump-1 protease (PUMP-1), or uterine metalloproteinase is an enzyme in humans that is encoded by the MMP-7 gene. <br /> |
The primary role of cleaved/activated MMP-7 is to break down extracellular matrix by degrading macromolecules including casein, type I, II, IV, and V gelatins, fibronectin, and proteoglycan. Most MMP's are secreted as inactive proproteins which are activated when cleaved by extracellular proteinases. The enzyme encoded by this gene degrades proteoglycans, fibronectin, elastin and casein and differs from most MMP family members in that it lacks a conserved C-terminal protein domain.</p> | The primary role of cleaved/activated MMP-7 is to break down extracellular matrix by degrading macromolecules including casein, type I, II, IV, and V gelatins, fibronectin, and proteoglycan. Most MMP's are secreted as inactive proproteins which are activated when cleaved by extracellular proteinases. The enzyme encoded by this gene degrades proteoglycans, fibronectin, elastin and casein and differs from most MMP family members in that it lacks a conserved C-terminal protein domain.</p> | ||
| - | <p class="STYLE33"> | + | <p class="STYLE33">MMP-7 cleaves collagen III/IV/V/IX/X/XI and proteoglycan indicating that MMP inhibitors can potentially be used in therapies that involved in inhibition tissue degradation, remodeling, anti-angiogenesis and inhibition of tumor invasion. The upregulation of MMP-7 is associated with many malignant tumors including esophagus, stomach, colon, liver, pancreas, and renal cell carcinomas. High MMP-7 expression facilitates cancer invasion and angiogenesis by degrading extracellular matrix macromolecules and connective tissues. Theses degradations are associated with many mechanisms including embryogenesis, postpartum uterine involution, tissue repair, angiogenesis, bone remodeling, arthritis, decubitus ulcer, and tumor metastasis/invasion. Benifiting from the characteristic of MMP-7, it may be used to predict the level of cancer.</p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">Most MMP's are secreted as inactive proproteins which are activated when cleaved by extracellular proteinases. The enzyme encoded by this gene degrades proteoglycans, fibronectin, elastin and casein and differs from most MMP family members in that it lacks a conserved C-terminal protein domain.</p> |
<p class="STYLE33"><strong>What is CheZ?</strong></p> | <p class="STYLE33"><strong>What is CheZ?</strong></p> | ||
| - | <p class="STYLE33"> | + | <p class="STYLE33">Bacterial chemotaxis results from the ability of flagellated bacteria to control the frequency of switching between smooth-swimming and tumbling episodes in response to changes in concentration of extracellular substances. High levels of phosphorylated CheY protein are the intracellular signal for inducing the tumbling mode of swimming. The CheZ protein has been shown to control the level of phosphorylated CheY by regulating its rate of dephosphorylation. In short, protein CheZ plays an important role in bacterial chemotaxis signal transduction pathway by accelerating the dephosphorylation of phosphorylated CheY (CheY-P).</p> |
<p class="STYLE33"><strong class="STYLE48">References</strong></p> | <p class="STYLE33"><strong class="STYLE48">References</strong></p> | ||
| - | <p class="STYLE33"> | + | <p class="STYLE33">[1] Blanco L P, Evans M L, Smith D R, et al. Diversity, biogenesis and function of microbial amyloids[J]. Trends in microbiology, 2012, 20(2): 66-73.</p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">[2] Barnhart M M, Chapman M R. Curli biogenesis and function[J]. Annual review of microbiology, 2006, 60: 131.</p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">[3] Chen A Y, Deng Z, Billings A N, et al. Synthesis and patterning of tunable multiscale materials with engineered cells[J]. Nature materials, 2014.</p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">[4] Daniel M C, Astruc D. Gold nanoparticles: assembly, supramolecular chemistry, quantum-size-related properties, and applications toward biology, catalysis, and nanotechnology[J]. Chemical reviews, 2004, 104(1): 293-346.</p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">[5] Patolsky F, Timko B P, Zheng G, et al. Nanowire-based nanoelectronic devices in the life sciences[J]. MRS bulletin, 2007, 32(02): 142-149.</p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">[6] Wilipedia</p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">[7] Yokoyama Y, Grünebach F, Schmidt SM, Heine A, H?ntschel M, Stevanovic S, Rammensee HG, Brossart P (2008). "Matrilysin (MMP-7) is a novel broadly expressed tumor antigen recognized by antigen-specific T cells". Clin. Cancer Res. 14 (17): 5503–11. </p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">[8] Edman K, Furber M, Hemsley P, Johansson C, Pairaudeau G, Petersen J, Stocks M, Tervo A, Ward A, Wells E, Wissler L (2011). "The discovery of MMP7 inhibitors exploiting a novel selectivity trigger". ChemMedChem 6 (5): 769–73.</p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">[9] Sanna M G, Simon M I. In vivo and in vitro characterization of Escherichia coli protein CheZ gain-and loss-of-function mutants[J]. Journal of bacteriology, 1996, 178(21): 6275-6280.</p></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 615: | Line 620: | ||
<p class="STYLE33"> [14] Majzik, A. et al. (2010). Functionalization of gold nanoparticles with amino acid, β-amyloid peptides and fragment. Colloids and Surfaces B: Biointerfaces, 81(1), 235-241.</p> | <p class="STYLE33"> [14] Majzik, A. et al. (2010). Functionalization of gold nanoparticles with amino acid, β-amyloid peptides and fragment. Colloids and Surfaces B: Biointerfaces, 81(1), 235-241.</p> | ||
<p class="STYLE33"> [15] Liu G, Wang J, Wunschel DS, Lin Y (2006) Electrochemical proteolytic beacon for detection of matrix metalloproteinase activities. J Am Chem Soc 128(38):12382–12383. doi:10.1021/ja0626638</p> | <p class="STYLE33"> [15] Liu G, Wang J, Wunschel DS, Lin Y (2006) Electrochemical proteolytic beacon for detection of matrix metalloproteinase activities. J Am Chem Soc 128(38):12382–12383. doi:10.1021/ja0626638</p> | ||
| - | <p class="STYLE33"> [16] | + | <p class="STYLE33"> [16] ?Hammer N D, Schmidt J C, Chapman M R. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization[J]. Proceedings of the National Academy of Sciences, 2007, 104(30): 12494-12499. </p> |
<p class="STYLE33"> [17] Chen A Y, Deng Z, Billings A N, et al. Synthesis and patterning of tunable multiscale materials with engineered cells[J]. Nature materials, 2014. </p> | <p class="STYLE33"> [17] Chen A Y, Deng Z, Billings A N, et al. Synthesis and patterning of tunable multiscale materials with engineered cells[J]. Nature materials, 2014. </p> | ||
<p class="STYLE33"> [18] Scheibel T, Parthasarathy R, Sawicki G, et al. Conducting nanowires built by controlled self-assembly of amyloid fibers and selective metal deposition[J]. Proceedings of the National Academy of Sciences, 2003, 100(8): 4527-4532.</p></td> | <p class="STYLE33"> [18] Scheibel T, Parthasarathy R, Sawicki G, et al. Conducting nanowires built by controlled self-assembly of amyloid fibers and selective metal deposition[J]. Proceedings of the National Academy of Sciences, 2003, 100(8): 4527-4532.</p></td> | ||
| Line 626: | Line 631: | ||
<tr> | <tr> | ||
<td bgcolor="#FFED97"><p class="STYLE79">Part 1. Controllable Expression of Curli fiber</p> | <td bgcolor="#FFED97"><p class="STYLE79">Part 1. Controllable Expression of Curli fiber</p> | ||
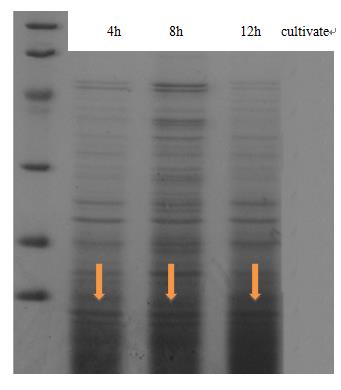
| - | <p class="STYLE33"> | + | <p class="STYLE33">Expression and secrete of CsgA, the major subunit of curli fiber, was carried out in <em>E coli.</em> trans-T1 transformed with CsgA, CsgB expression cassette (BBa_K1361007 docked into pSB1C3) and curli-secrete device (BBa_K1361005 docked into pSB3K3). We tracked cell growth and total quantities of secrete protein in culture. Samples were taken from culture and Bradford protein assay was adopted after centrifuging and sterilization filtered through 0.22μm filter membrane. Meanwhile, another sample taken at 2-hour interval was wall-broken and examined by SDS-PAGE, which confirm the expression of CsgA-his.</p> |
<table width="100%" border="0" cellspacing="0" cellpadding="0"> | <table width="100%" border="0" cellspacing="0" cellpadding="0"> | ||
<tr bgcolor="#FFED97"> | <tr bgcolor="#FFED97"> | ||
| Line 634: | Line 639: | ||
<td class="STYLE33">Fig 1. The result of SDS-PAGE shows that CsgA-his has been constitutive expressed in cell. Inducating that BBa_K1361007 works as anticipation.</td> | <td class="STYLE33">Fig 1. The result of SDS-PAGE shows that CsgA-his has been constitutive expressed in cell. Inducating that BBa_K1361007 works as anticipation.</td> | ||
</tr> | </tr> | ||
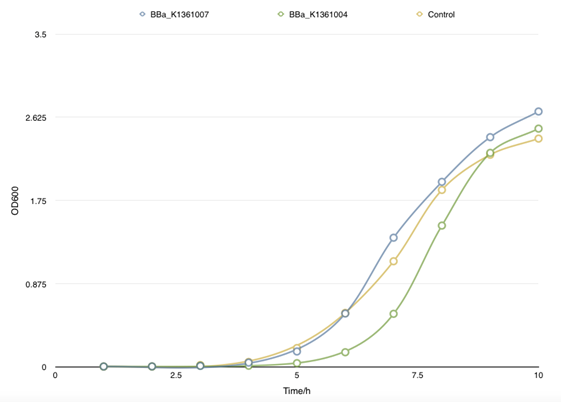
| - | </table> <p class="STYLE33"> | + | </table> <p class="STYLE33">In order to find the proper promoter strength to control the concentration of CsgA monomer, Cell growth curve was drew to compare the CsgA secrete and self- aggregation level when adapted with a strong promoter BBa_J23100 and a weaker one BBa_J23104. </p> |
<table width="100%" border="0" cellspacing="0" cellpadding="0"> | <table width="100%" border="0" cellspacing="0" cellpadding="0"> | ||
<tr> | <tr> | ||
| Line 652: | Line 657: | ||
</table> | </table> | ||
<p class="STYLE33"> </p> | <p class="STYLE33"> </p> | ||
| - | <p class="STYLE33"> | + | <p class="STYLE33">However, there is no obvious divergence of the concentration of matrix soluble protein between control samples (with empty plasmids) and cells that had been transformed with both BBa_K1361007/BBa_K1361004 (Curli Fiber generator under the control of Pbad promoter with CsgA modified by His tag) and BBa_K1361005 (the outer membrane secrete device for curli fiber). Coupled with the failure of CsgA-his purification from cell culture, suggesting that the outer membrane channel protein CsgG and its assist proteins CsgE, F we using wasn’t functioning and may get mutation.</p> |
<p class="STYLE33"><strong>Part 2. Congo Red (CR) Assay to Determine the Degree of Curli Fiber Polymerization</strong></p> | <p class="STYLE33"><strong>Part 2. Congo Red (CR) Assay to Determine the Degree of Curli Fiber Polymerization</strong></p> | ||
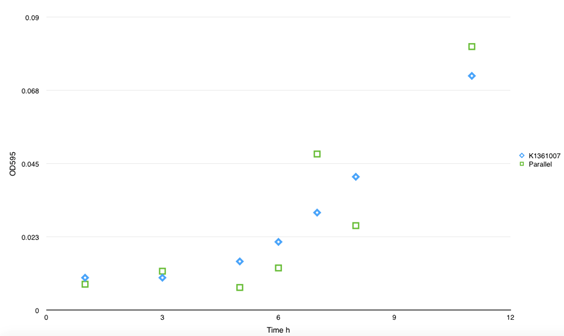
| - | <p class="STYLE33"> | + | <p class="STYLE33">To examine our controlling force on curli fiber inductively aggregation, we measured the curli fiber polymerization properties after L-arabinose inducing by CR assay. The experiments showed that adding of the inducer seemed not greatly affect the curli fiber assembly. Regardless of induction, there would be decay at A480 within 10min after transferring cells into static condition.</p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">Speculating the non-function of outer membrane secrete system, we restructured the out membrane channel CsgEFG and send it to iGEM Headquarter (BBa_K1361005), while using it to process further characterization. Result of which will be demonstrated in our presentation. </p> |
<table width="100%" border="0" cellspacing="0" cellpadding="0"> | <table width="100%" border="0" cellspacing="0" cellpadding="0"> | ||
<tr> | <tr> | ||
| Line 661: | Line 666: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td bgcolor="#FFED97"><span class="STYLE33">Fig 4. The polymerization rate of curli fiber was determined by CR assay. | + | <td bgcolor="#FFED97"><span class="STYLE33">Fig 4. The polymerization rate of curli fiber was determined by CR assay.? After 1μM L- arabinose inducing at 0min, culture medium was subpackaged into glass tubes and cultivates under 37C, static. Samples were taken to filter and suspended in 700μl 1XPBS; 300μl of cell suspension was used for OD600 cell number normalization, and the remaining 400μl combined with 5X Congo Red (CR) for a final concentration of 20μg/ml CR, and incubated for 5min at RT. The cells and curli with bound CR were spun down at 15,000-x g for 5min, and 300μl supernatant was removed for CR quantification. Concentration of CR in supernatant was quantified by absorbance at 480nm. The amount of CR bound by cells and curli were quantified by subtracting the Ab480nm of supernatant from Ab480nm of 20μg/ml CR. </span></td> |
</tr> | </tr> | ||
</table> | </table> | ||
| Line 687: | Line 692: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td bgcolor="#FFED97"><p class="STYLE33"> | + | <td bgcolor="#FFED97"><p class="STYLE33">We believe the enlightening idea of the transformation from bio-signal to electrical signal will exert far-reaching influence on the future society for the convenience and the help of highly efficient computing and analyzing capacity of computers. Our project is born ignited attributed to the belief, and as a result, we have devoted ourselves to the theories and experiments constructing to the final project. However, some work is expected to be done to perfect the project in the future due to the limited time we have during the competition.</p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">Firstly, no ideal method have been found to reverse the electroconductivity when the concentration of the seducer is lower than the threshold value. Although we have paid great effort on searching information on the problem and assumed that enzymolysis and the motivation of bacterial flagella can destroy the nanowire composed by CsgA, no further experiments can be carried on for the lack of time and grounded references.</p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">Secondly, the linker used in the experiment of the detection of MMP-7 is obtained semiempirically. Thus, this linker will potentially and probably effect the protein folding of CsgA and the substrate polypeptide, exert negative influence on the aggregation of CsgA and the enzymolysis of the polypeptide by MMP-7. Consequently, an optimal linker may be found through future theoretical calculation and experiments to lead to better results.</p> |
| - | <p class="STYLE33"> | + | <p class="STYLE33">Finally, the application of the project will be considered in the future. This project is endowed with innovative ideas and constituted of various sub-modules, but the experiments only prove that the system can be a great practical device to detect the existence of protease(MMP-7). We believe the ideas embedded in the project can ignite our ideas of the novel application of biosensors related to pharmacy, detection of organics, medicine treatment, etc.</p></td> |
</tr> | </tr> | ||
</table></td> | </table></td> | ||
| Line 701: | Line 706: | ||
<tr> | <tr> | ||
<td width="19%" bgcolor="#3A2108"><a href="http://www.tju.edu.cn/"><img src="https://static.igem.org/mediawiki/2014/0/01/Tianjin-school.png" width="175" height="163" border="0" /></a></td> | <td width="19%" bgcolor="#3A2108"><a href="http://www.tju.edu.cn/"><img src="https://static.igem.org/mediawiki/2014/0/01/Tianjin-school.png" width="175" height="163" border="0" /></a></td> | ||
| - | <td width="81%" bgcolor="#3A2108"><p><span class="STYLE61"> | + | <td width="81%" bgcolor="#3A2108"><p><span class="STYLE61">Tianjin University,Tianjin, China<br /> |
| - | + | Email:michaelss@tju.edu.cn</span></p> | |
| - | + | ||
| - | + | ||
| - | + | ||
<p> </p></td> | <p> </p></td> | ||
</tr> | </tr> | ||
Revision as of 17:50, 17 October 2014
|
|
Welcome to Team Tianjin! |
 |
|
 "
"