Team:Saarland/Modeling
From 2014.igem.org
Modelling:
The hyaluronan synthase of the naked mole rat is in possession of 7 putative transmembrane domains and one large cytoplasmic loop which is supposed to be the catalytic center for HA synthesis. As there are no crystal structures of hyaluronan synthases, the corresponding protein modelling was performed using the template based RaptorX web server (Källberg et al., 2012; Ma et al., 2013; Peng and Xu, 2011; Peng and Xu, 2011b). It is shown in figure 1.
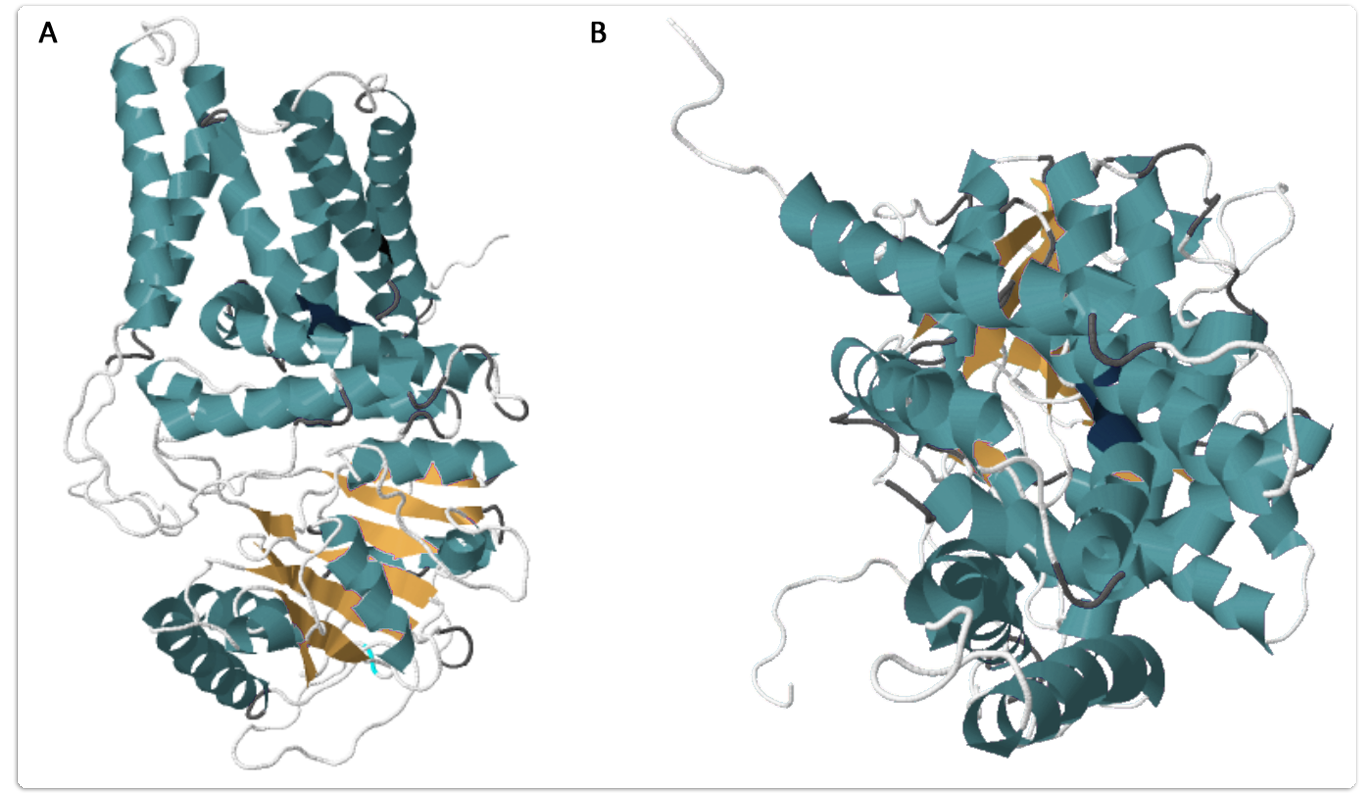
In contrast to the highly conserved catalytic domain of hyaluronan synthases in other vertebrates the naked mole rat´s HaS features two unique changes in the amino acid sequence as two asparagines have been substituted by serines. This might be the cause for the raised activity of the naked mole rat´s Has. There are two different models described for HA transport across the membrane. It´s either possible that there is a specific transporter for oligosaccharides mediating HA secretion into the medium, or that the multi transmembrane domains of the hyaluronan synthase themselves form a channel for the secretion of the nascent HA chain. Based on the protein model we think the more likely model for HA secretion across the membrane is the channel.
References:
Källberg, M., Wang, H., Wang, S., Peng, J., Wang, Z., Lu, H., and Xu, J. (2012). Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 7, 1511–1522.
Ma, J., Wang, S., Zhao, F., and Xu, J. (2013). Protein threading using context-specific alignment potential. Bioinformatics 29, i257–i265.
Peng, J., and Xu, J. (2011a). A multiple-template approach to protein threading. Proteins 79, 1930–1939.
Peng, J., and Xu, J. (2011b). Raptorx: Exploiting structure information for protein alignment by statistical inference. Proteins Struct. Funct. Bioinforma. 79, 161–171.
 "
"



Impressum/Copyright