Team:Saarland/Methods
From 2014.igem.org
Methods
The following section lists the used methods in the presented iGEM Project:

Skip to a chapter by clicking the corresponding marking above
1. Cloning
1.1 Restriction and phosphatase treatment of plasmid DNA
Originally found in bacteria, endonucleases belong to a class of enzymes capable of cleaving the phosphodiester bond of DNA molecules by hydrolysis. Depending on the type they recognize site specific base sequences of 4, 6 or 8 bases respectively, which are often palindromic. Treatment of different DNA molecules with the same endonucleases allows the combination of the resulting DNA fragments in a following ligation reaction.
The treatment of DNA fragments with phosphatase is beneficial if the restriction is performed with only one restriction enzyme. It can also be beneficial if the restriction with two different restriction enzymes is unbalanced and single cut plasmid DNA is left. This applies in particular to the treatment of plasmid DNA with phosphatase. As the nomenclature indicates the phosphatase cleaves the phosphate group at the 5´terminus of DNA fragments and thereby minimizes the background of mono ligated plasmid DNA. Consequently the amount of transformants with insert DNA in the plasmid construct is considerably increased.
The following components were used for a basic restriction mixture:
| 10 x FastDigest Buffer | 3 µl |
| Restriction Enzyme 1 | 10 U |
| Restriction Enzyme 2 | 10 U |
| FastAP | 1 U |
| Plasmid | approx. 1 µg |
| ddH2O | to 30 µl |
The mixture was incubated for 10 min at 37 °C. The success of the restriction was subsequently checked in an agarose gel electrophoresis.
1.2 Restriction of PCR products:
Corresponding restriction sites were added in a PCR reaction at the 5´and 3´termini of insert DNA via modified primers. After gelextraction the following components were used for a basic restriction mixure.
| 10 x FastDigest Buffer | 3 µl |
| Restriction Enzyme 1 | 10 U |
| Restriction Enzyme 2 | 10 U |
| PCR Product | 25 µl |
| ddH2O | to 30 µl |
The mixture was incubated for 10 min at 37 °C followed by purification via QIAquick Gel Extraction Kit (Qiagen).
1.3 Polymerase chain reaction (PCR)
The PCR is a standard procedure in molecular biology that enables the amplification of distinct DNA sequences in vitro. Short flanking DNA oligomers define the DNA sequence which is to be amplified. The amplification process itself is performed by a polymerizing enzyme also referred to as DNA polymerase. Since its invention in 1983 numerous variations have been established for different applications.
1.3.1 Amplification of genomic DNA:
The following components were used for the basic amplification of genomic DNA of Bacillus megaterium:
| 2x Q5 Master Mix | 10 µl |
| 10 µM Forward | 1 µl |
| 10 µM Reverse Primer | 1 µl |
| Genomic DNA | 100 ng |
| ddH2O | to 20 µl |
The following speed cycle program was used:
| Step | Temperature [°C] | Time [s] |
|---|---|---|
| 1. Initial Denaturation | 98 | 30 |
| 2. Denaturation | 98 | 15 |
| 3. Annealing | 60 | 15 |
| 4. Extention | 75 | 20/kb |
| 5. Final Extention | 72 | 120 |
| 6. storage | 4 | for ever |
 30 cycles
30 cycles
Presence, absence and size of amplification products were determined by agarose gel electrophoresis.
1.3.2 Splicing by overlapping extension (SOE)-PCR:
The SOE-PCR is a common variation of the standard PCR reaction and primarily used for the generation of fusion genes. The name refers to the special set of primers. Their single stranded endings are complementarily overlapping with the DNA sequences of the DNA fragments to be fused. Figure 1 shows the flowchart of the SOE-PCR for the generation of a hyaluronan synthase (has2) - green fluorescent protein (gfp) fusion gene.

The following components were used for the first standard PCR reactions:
| 2x Q5 Master Mix | 10 µl |
| 10 µM Forward Primer a or c | 1 µl |
| 10 µM Reverse Primer b or d | 1 µl |
| Plasmid DNA | 10 ng |
| ddH2O | to 20 µl |
The following components were used for the second standard PCR reaction:
| 2x Q5 Master Mix | 10 µl |
| 10 µM Forward Primer a | 1 µl |
| 10 µM Reverse Primer d | 1 µl |
| Amplification Product 1 and 2 | 1:1 Molar Ratio (approx. 10 ng) |
| ddH2O | to 20 µl |
The following speed cycle program was used:
| Step | Temperature [°C] | Time [s] |
|---|---|---|
| 1. Initial Denaturation | 98 | 30 |
| 2. Denaturation | 98 | 15 |
| 3. Annealing | 60 | 15 |
| 4. Extention | 75 | 20/kb |
| 5. Final Extention | 72 | 120 |
| 6. storage | 4 | for ever |
 30 cycles
30 cycles
Presence, absence and size of amplification products were determined by agarose gel electrophoresis.
1.3.3 Colony PCR:
After transformation of competent E. coli cells the colony PCR is a popular high-throughput technique in molecular biology for the quick identification of the presence, absence or orientation of insert DNA in plasmid constructs, respectively. An extended heating step at the beginning of the PCR reaction results in cell lysis and release of plasmid DNA which serves as template for the following amplification with insert specific primers. Time consuming DNA isolations and restrictions of multiple colonies are superfluous.
The following components were used for the second standard PCR reaction:
| 10x ThermoPol Reaction Buffer | 2 µl |
| 10 mM dNTP Mix | 0,4 µl |
| 10 µM Forward Primer | 1 µl |
| 10 µM Reverse Primer | 1 µl |
| Taq DNA Polymerase | ½ U |
| ddH2O | to 20 µl |
| colony | Resuspend |
The following speed cycle program was used:
| Step | Temperature [°C] | Time [s] |
|---|---|---|
| 1. Initial Denaturation | 95 | 180 |
| 2. Denaturation | 95 | 30 |
| 3. Annealing | 60 | 30 |
| 4. Extention | 68 | 60/kb |
| 5. Final Extention | 68 | 300 |
| 6. storage | 4 | for ever |
 34 cycles
34 cycles
Presence, absence and size of amplification products were determined by agarose gel electrophoresis.
1.3.4 Quick Change (QC)-PCR:
The QC-PCR is a common variation of the standard PCR and useful for site-directed mutagenesis of plasmid DNA. The special design of mutagenic primers allows the insertion or deletion of whole sequences, respectively, as well as the substitution of single nucleotides for applications such as restriction site mutagenesis or amino acid exchange. Figure 2 shows the experimental setup for a QC-PCR.
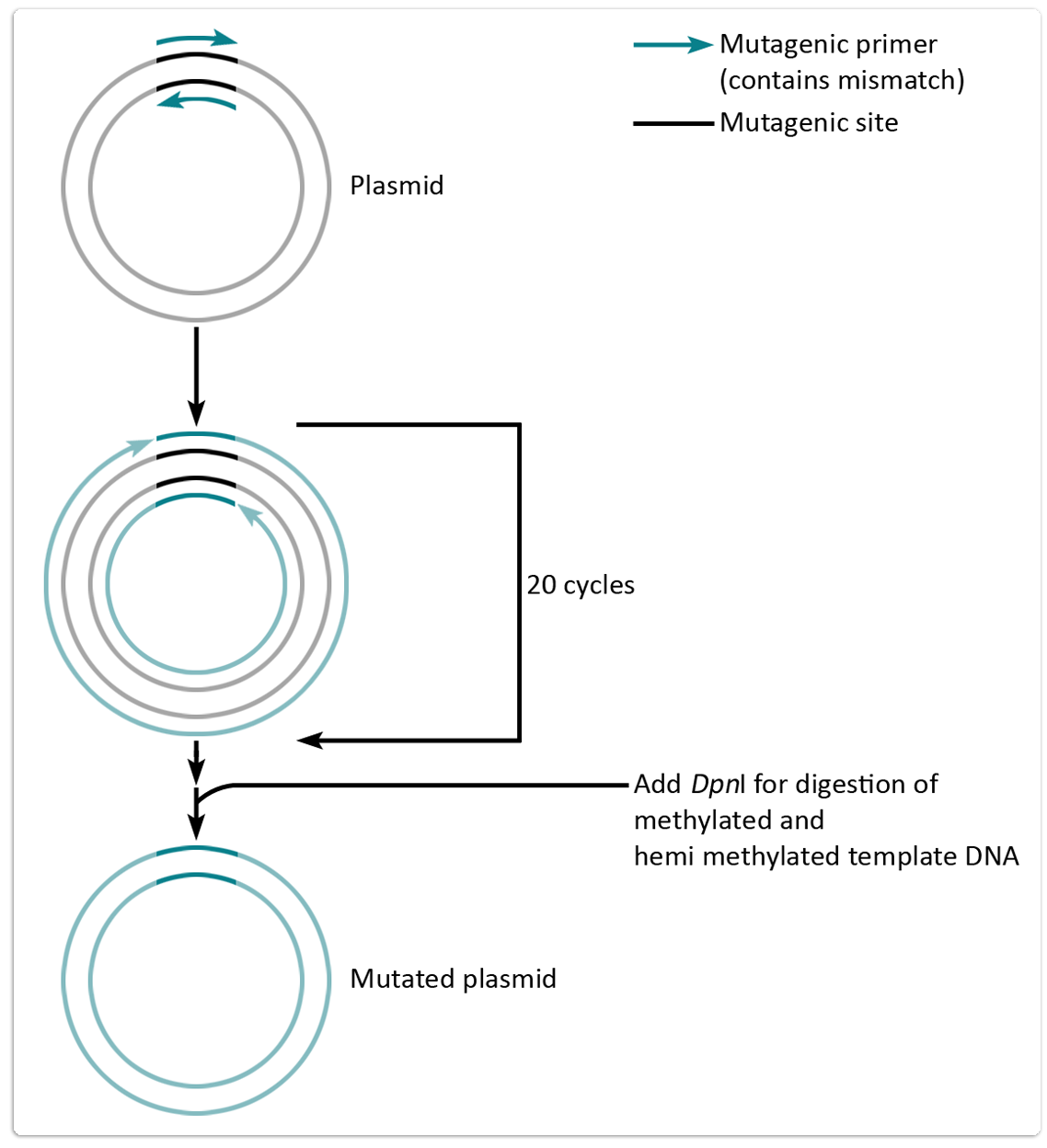
The following components were used for the standard PCR reactions:
| 2x Q5 Master Mix | 25 µl |
| 10 µM Forward QC Primer | 2,5 µl |
| 10 µM Reverse QC Primer | 2,5 µl |
| Plasmid DNA | 10 ng |
| ddH2O | to 50 µl |
The following speed cycle program was used:
| Step | Temperature [°C] | Time [s] |
|---|---|---|
| 1. Initial Denaturation | 95 | 30 |
| 2. Denaturation | 98 | 15 |
| 3. Annealing | 60 | 35 |
| 4. Extention | 72 | 20/kb |
| 5. Final Extention | 72 | 300 |
| 6. storage | 4 | for ever |
 20 cycles
20 cycles
The following components were used for digestion and removal of the template plasmid DNA :
| 10 x FastDigest Buffer | 5 µl |
| DpnI | 1 U |
| QC-PCR Batch | 40 µl |
| ddH2O | to 50 µl |
Presence, absence and size of amplification products were determined by agarose gel electrophoresis after digestion with DpnI.
1.4 Isolation of plasmid DNA
The isolation of plasmid DNA was performed with a QIAprep® Spin Miniprep Kit (Qiagen). The provided instructions were followed precisely.
1.5 Isolation of genomic DNA of Bacillus megaterium:
The isolation of genomic DNA of Bacillus megaterium was performed with a NexttecTM 1-Step DNA Isolation Kit for Bacteria. The provided instructions were followed precisely.
1.6 Measurement of DNA concentration:
The measurement of DNA concentration after plasmid DNA isolation was performed with the UV2100 UV/Vis Recording Spectrophotometer (Shimadzu). The spectrum was analysed between wavelengths of 300 nm and 200 nm. The level of DNA purity was assessed with the q-value of A260 /A280.
1.7 Agarose gel electrophoresis (AGE)
The agarose gel electrophoresis is a standard method in molecular biology primarily used for separation of DNA or RNA molecules of varying lengths, respectively. The separation effect is due to the filtering properties of the three dimensional agarose gel matrix. After applying an electric field it enables short negatively charged DNA molecules to travel faster towards the positive anode than large negatively charged DNA molecules. As a consequence the distribution pattern of DNA molecules is inversely proportional to their molecular weights. DNA fragments with identical lengths form bands inside the gel. After separation and staining with an appropriate dye (e.g. ethidium bromide) the DNA bands can be visualized under ultra violet light.
The agarose gel electrophoresis was performed with prestained (peqGREEN) 1 % agarose gels (w/v) in 0.5 x TBE buffer. The 2-Log DNA Ladder (NEB) was used as indicator for DNA size. 120 V were applied for 50 min for appropriate separation.
1.8 Gel extraction and purification of DNA fragments
Both procedures were performed with the QIAquick Gel Extraction Kit (Qiagen). The provided instructions were followed precisely with the exception that the elution step was performed with 25 µl preheated ddH2O.
1.9 Ligation of DNA fragments
In the ligation process selected DNA fragments such as insert DNA and plasmid constructs are enzymatically linked to form recombinant DNA molecules. The phosphodiester bond is formed between 3`hydroxyl groups and 5`phosphoryl groups of terminal nucleotides. The energy for the reaction is provided by addition of the co-factor ATP.
The following components were used for the basic ligation mixture:
| 10 x T4 Ligation Buffer | 2 µl |
| Insert DNA and Plasmid Construct | 3:1 Molar Ratio (approx. 50 ng) |
| T4 DNA Ligase | 1 U |
| ddH2O | to 20 µl |
The ligation reaction was incubated for 10 min at 22 °C. Heat inactivation was performed for 10 min at 65°C. Success of the ligation reaction was checked via agarose gel electrophoresis with 5 µl of the ligation batch.
1.10 Dialysis of the ligation mixture
Dialysis of the ligation mixture was performed on MF-Membrane Filter Type VSWP02500 (Millipore) in 10 % glycerol for 1h at RT. Transformation of competent E. coli cells was subsequently performed with 5 µl of the dialyzed ligation batch.
2. Transformation and cultivation of E. coli and B. megaterium
2.1 Measurement of cell density
Measurement of cell density was regularly performed after inoculation of cultures with 1 ml samples. Fresh LB medium or TB medium was used as reference for measurement of E. coli or B. megaterium density, respectively. The measurement itself was performed with a Novaspec visible spectrophotometer at a wavelength of 600 nm.
2.2 Preparation of electro competent E. coli cells
E. coli cells belong to the class of Gram-negative bacteria and are therefore not naturally competent for absorption of DNA molecules. For a successful transformation with plasmid DNA, E. coli cells have to be made competent artificially. The following protocol was used for preparation of electro competent E. coli cells with high transformation efficiency of 2 x 108 cfu/µg pUC19 DNA:
2x 400 ml LB medium in 2000 ml Erlenmeyer flasks were inoculated with one hundredth of a fresh overnight culture of the E. coli cloning strain TOP10F´. The main cultures were incubated at 37 °C and 180 rpm until a cell density of A600=0.4. The measurement of the cell density was regularly performed over a period of time of approximately 3 hours. The flasks were subsequently chilled on ice for 30 minutes. Afterwards each culture was centrifuged at 4000 g and 4 °C for 10 min. The supernatant was discarded and each pellet resuspended in 400 ml of ice cold, sterile water. Another centrifugation step followed. The supernatant was discarded and each pellet resuspended in 200 ml of ice cold, sterile water. The centrifugation step was repeated. The supernatant was discarded and each pellet resuspended in 100 ml of ice cold, sterile water. Another centrifugation step followed. The supernatant was discarded and each pellet resuspended in 50 ml of ice cold, sterile water. The centrifugation step was repeated. The supernatant was discarded and each pellet resuspended in 20 ml of ice cold, sterile 10 % glycerol. Another centrifugation step followed. The supernatant was discarded and each pellet resuspended in 10 ml of ice cold, sterile 10 % glycerol. The centrifugation step was repeated. The supernatant was discarded and each pellet resuspended in 2 ml of ice cold, sterile 10 % glycerol. The electro competent E. coli cells were immediately frozen in aliquots of 50 µl and kept at -70 °C until reconstitution.
2.3 Transformation of electro competent E.coli cells:
In the electroporation process, the membrane of E. coli cells is made permeable by applying a high voltage over a short period of time. Frozen electro competent E.coli cells were slowly thawed on ice and mixed with up to 5 µl of prechilled, dialyzed DNA. After careful resuspension the transformation mixture was incubated for 5 minutes on ice and subsequently filled into a prechilled electroporation cuvette (gap width 0.1 cm). The transformation was performed via electroporation with the Easyject Prima (Equibio) at 1800 V. The transformation mixture was immediately mixed with 500 µl of preheated SOC medium and incubated at 37 °C and 700 rpm for 1 h. Different dilutions of the transformation mixture were grown on LB agar plates with an appropriate selection medium at 37 °C over night. Presence and size of insert DNA in plasmid constructs of transformants were checked by colony PCR.
2.4 Cultivation of E. coli cells
A 300 ml Erlenmeyer flask was filled with 50 ml of fresh LB medium and 50 µl of an ampicillin stock solution (100 mg/ml). The LB medium is inoculated with an E. coli colony that has been positively tested for insert DNA by colony PCR. The resulting culture was incubated at 37 °C and 180 rpm over night.
2.5 Preparation of B. megaterium protoplasts
B. megaterium belongs to the class of gram positive bacteria. Thus it is naturally competent for absorption of DNA. However, the thick surrounding peptidoglycane layer prevents the absorption of DNA and has to be degraded. The following protocol was used for preparation of protoplasts.
50 ml LB medium in a 300 ml baffeled flask was inoculated with one hundredth of a fresh overnight culture of the B. megaterium strain MS941. The main culture was incubated at 37 °C and 180 rpm until a cell density of A600=1.0. The measurement of the cell density was regularly performed over a period of time of approximately 4 hours. The culture was transferred to a 50 ml Falcon tube and centrifuged at 1280 g and 4 °C for 15 min. The supernatant was discarded, the pellet was resuspended in 5 ml of fresh SMMP and transferred to a new 15 ml Falcon tube. The protoplast formation was initiated by addition of 100 µl of lysozyme solution (10 mg/ml in SMMP). The mixture was incubated at 37 °C without shaking until 80-100 % of the cells showed the typical globular morphology of protoplasts. The progress of the reaction was checked under a microscope every 10 min. If the formation of protoplasts has not been initiated, another 100 µl of the lysozyme solution should be added. After successful protoplast formation, the mixture was centrifuged at 1280 g and RT for 10 min. The supernatant was carefully removed and the pellet was washed with 5 ml of SMMP. Both, centrifugation and washing step were repeated. Subsequently 750 µl of sterile 87 % glycerol were added to the protoplast suspension and carefully mixed by tilting the tube. Finally the protoplasts were frozen in 500 µl aliquots and kept at-70 °C until reconstitution.
2.6 Transformation of B. megaterium protoplasts
A 15 ml Falcon tube was prepared and filled with 1.5 ml of PEG-P which serves as a carrier matrix for the DNA. The protoplast suspension was slowly thawed on ice and carefully mixed with 5-10 µg of DNA. With a micropipette in circling motions the protoplast-DNA-solution was subsequently pulled vertically through the PEG-P. The resulting mixture was mixed by tilting the tube and was incubated for 2 min at RT. Afterwards the mixture was resuspended with 5 ml of SMMP and centrifuged at 1280 g and RT for 10 min. The supernatant was immediately removed, the pellet resuspended in 500 µl SMMP and transferred into a sterile 1.5 ml reaction vessel. The regeneration of the cell wall was triggered in two steps. The first incubation step was performed at 30 °C without rotations, while the second one was performed at 30°C and 300 rpm. During these 90 min incubation time, 3.5 ml of CR5 overlay agar were prepared in a 15 ml Falcon tube and kept warm in a water bath at a temperature of 42 °C. After incubation the transformation mixture was added to the overlay agar and carefully mixed by tilting the 15 ml Falcon tube. The agar was poured onto LB plates with an appropriate selection medium. The plates were incubated over night at 37 °C. The next day single colonies were transferred to fresh LB plates with selection medium and incubated again over night at 37 °C. To avoid contamination, the following day single colonies were identified via microscopy as representatives of B. megaterium.
2.7 Cultivation of B. megaterium cells
A 300 ml Erlenmeyer flask was filled with 50 ml of LB medium and 50 µl of a tetracycline stem solution (10 mg/ml). Transformed B. megaterium strains were taken from the corresponding glycerol stock and put into the Erlenmeyer flask. This preculture was incubated at 37 °C and 180 rpm overnight. The next day a mix of 45 ml fresh TB medium, 5 ml 10 x KPP buffer and 50 µl tetracycline stem solution was inoculated in a 300 ml baffled flask with one hundredth of the fresh overnight culture of the B. magisterium and incubated at 30 °C and 180 rpm until a cell density density of A600= 0,4. Protein expression was induced by addition of 1 ml of a xylose solution (250 mg/ml) followed by further incubation at 30 °C and 180 rpm.
3. Protein expression analysis:
3.1 Sample preparation for SDS Page
Protein expression levels were checked after 48 h of cultivation. Cell density was adjusted to an A600 nm = 3. Cell pellets were resuspended in 30 μl of lysis buffer and incubated for 30 min at 37 °C and 1,000 rpm. Vortexing every 10 minutes should increase cell lysis. Separation of insoluble fraction (pellet) from the soluble fraction (supernatant) was performed by centrifugation for 30 min at 4 °C and 13,000 rpm. 24 µl of the supernatant (containing soluble proteins) were mixed with 6 µl of SDS sample buffer. Subsequently, the supernatant was completely removed. The pellet fraction was resuspended in 30 ml of 8 % urea (w/v) and centrifuged for 30 min at 4 °C and 13,000 rpm. 24 µl of the supernatant (containing insoluble proteins) were mixed with 6 µl of SDS sample buffer. Each sample was heated for 5 min at 95 °C. 7 µl of each sample were used for protein separation by SDS-PAGE.
3.2 SDS-PAGE:
The samples were separated using a 12 % separating gel and a 4% stacking gel. 7 µl of each sample were loaded onto the gel. The gel electrophoresis ran for approximately 40 min at 20 mA.
3.3 Western Blot:
The used procedure is a semi-dry blotting procedure.
The previously separated samples were transferred to a nitrocellulose membrane. The gel was put under voltage (20 V; intensity: 0,4 A; power: 8 W) during 40 min. Afterwards the membrane was incubated in blocking buffer for 60 min on a shaking device at room temperature. The nitocellulose membrane was then incubated in primary anti-GFP antibody (rabbit) overnight at 4 °C on a shaking device. The membrane was rinsed with water and washed with PBS-T (3x 5 min) to eliminate the unbound antibodies. The blot was incubated with secondary anti-rabbit peroxidase antibodies for 60 min at room temperature on a shaking device. After this step the washing procedure was repeated. Subsequently, the nitrocellulose membrane was put in a transparent foil and residual liquid was removed. The Western Lightning Enhanced Chemiluminescence substrates were mixed in equal parts and evenly spread on the membrane. After an incubation of 10 s the substrates were removed. The blot was developed on an x-ray film with developing and fixating solution in the dark room.
3.4 Sample preparation for fluorescence microscopy:
Localisation studies of the GFP tagged hyaluronan synthase (Has2) were carried out after 24 h and 48 h of cultivating the corresponding B. megaterium strains. 1 ml sample was centrifuged for 1 min at 11000 g and the cell pellet was washed with 1 ml of ddH2O. 10 µl of the cell suspension were put on a microscope slide an air dried.
4. Confirmation of hyaluronic acid production by rheometry
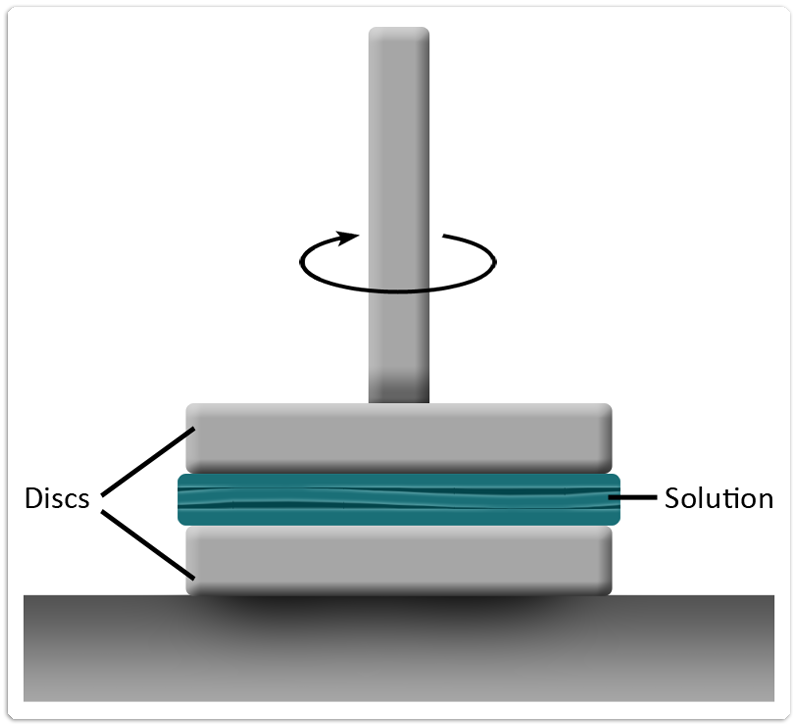
Since hyaluronic acid is very viscous and 1 g of it can bind 6 litres of water, the viscosity of the medium should rise significantly (at least by a factor of 10-100) when hyaluronic acid is synthesised. Viscosity measurements were carried out using a parallel discs rheometer.
Rheometers are generally used for measuring the (complex) viscosity of materials dependent on quantities like temperature, time or shear velocity. As shown in figure 3, a parallel disc rheometer is composed of two horizontal discs. The bottom disc is fixed and the upper disc is connected to anelectric motor. For our experiments, 1 ml of sample was used.
With a distance of 20 nm to the lower disc, the upper disc is set into rotation and the torque needed to sustain this motion is measured. Based on the torque, shear stress τ in and thus the viscosity η of the fluid can be calculated:
Because hyaluronic acid is a pseudoplastic fluid, meaning that its viscosity decreases with rising shear forces, the measurement is done over a range of shear forces and should result in a characteristic curve like the one shown in figure 4 if hyaluronic acid is present within the solution.
 "
"




Impressum/Copyright