Team:Paris Saclay/Modeling
From 2014.igem.org
m (→Modelling) |
m (→Modelling) |
||
| Line 9: | Line 9: | ||
At this step of the wiki, the attentive reader has certainly understood that the goal of our project is to raise up ethical questions by creating an agar lemon covered with bacteria - ''E.coli'' in practice. But, before starting more investigations and experimentations, there is a real need to ensure that our project is feasible. | At this step of the wiki, the attentive reader has certainly understood that the goal of our project is to raise up ethical questions by creating an agar lemon covered with bacteria - ''E.coli'' in practice. But, before starting more investigations and experimentations, there is a real need to ensure that our project is feasible. | ||
| - | For this reason, we think | + | For this reason, we think modelling is so much important in an iGEM project. |
| Line 33: | Line 33: | ||
''(* Firstly, we wonder if bacteria could live and develop them-selves correctly in the agar media. | ''(* Firstly, we wonder if bacteria could live and develop them-selves correctly in the agar media. | ||
| - | Actually, | + | Actually, ultimately we hope to create our lemon with the help of a 3D-printer which would use a mix of melted agar media and bacterial solution. As the first ingredient for bacterial growth is oxygen, we decided to model the [https://2014.igem.org/Team:Paris_Saclay/Modeling/oxygen_diffusion oxygen diffusion] in an agar medium.)'' |
| Line 41: | Line 41: | ||
* Now that we know if bacteria could develop in agar or not, a natural issue is to predict the [https://2014.igem.org/Team:Paris_Saclay/Modeling/bacterial_Growth bacterial growth]. | * Now that we know if bacteria could develop in agar or not, a natural issue is to predict the [https://2014.igem.org/Team:Paris_Saclay/Modeling/bacterial_Growth bacterial growth]. | ||
| - | Without spoiling, we can just say that the previous model | + | Without spoiling, we can just say that the previous model confirms the intuition we could have : while oxygen can't easily move into the medium, the growth on the surface area is sufficiently more important than in the medium to neglect the first one. More precisely, our bacteria is colorful and the surface will be entirely covered before internal bacteria have time to develop enough to be visible. Finaly, when we could hope to observe internal bacteria due to their color, the surface bacteria have already done an opaque layer. |
| - | So, this part aims | + | So, this part aims at predicting over time the bacterial population growth on an ellipsoidal object - a fake lemon in practice -. |
| - | More precisely, we have | + | More precisely, we have to determine the initial proportion of bacteria threshold from which we are sure that the population will never extinct...if this threshold exists ! |
---- | ---- | ||
Revision as of 23:58, 15 October 2014

Modelling
comment : mettre les points en + gros ou des trucs en gras car ça fait trop bloc Je pense que t'auras du mal à le faire. Je propose qu'on mettre une image en miniature (pas trop quand même) assez representative des parties. Par exemple votre dernier graphe de bacterial growth.
At this step of the wiki, the attentive reader has certainly understood that the goal of our project is to raise up ethical questions by creating an agar lemon covered with bacteria - E.coli in practice. But, before starting more investigations and experimentations, there is a real need to ensure that our project is feasible.
For this reason, we think modelling is so much important in an iGEM project.
 The realisation of our lemon requires introduction of bacteria in an agar gel. This poses several technical problems:
The realisation of our lemon requires introduction of bacteria in an agar gel. This poses several technical problems:
- bacteria need access to oxygen for respiration.
- We want our bacteria produce a "lemon coloration-like" in order to have a crust on the surface.
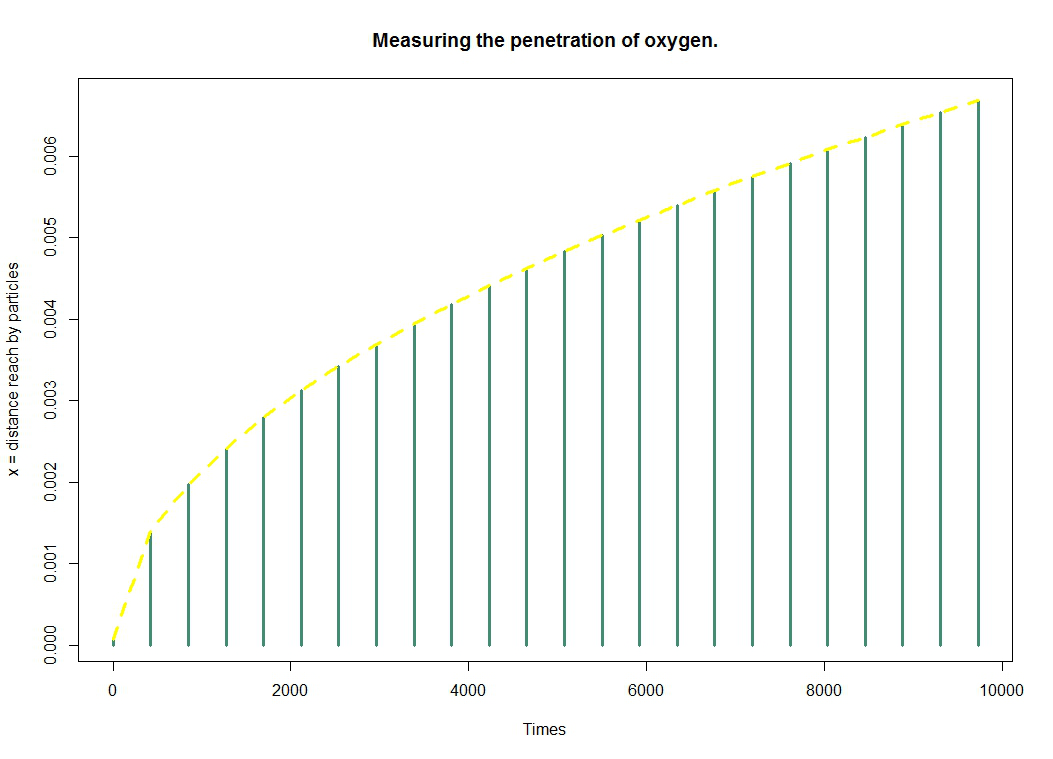
We will make a compromise to solve these two problems. The first one is easy to solve because E.coli can live in an anaerobic environment. Thus, this, combined with the ease of access of oxygen, we solve the second problem by reacting our E.coli only in contacts with oxygen. Since we just want a crust, it is not necessary that all our bacteria react. Hence the necessity to estimate the penetration of oxygen into the gel.
We invite you to discover how we model the oxygen diffusion.
A enlever!
(* Firstly, we wonder if bacteria could live and develop them-selves correctly in the agar media.
Actually, ultimately we hope to create our lemon with the help of a 3D-printer which would use a mix of melted agar media and bacterial solution. As the first ingredient for bacterial growth is oxygen, we decided to model the oxygen diffusion in an agar medium.)
- Now that we know if bacteria could develop in agar or not, a natural issue is to predict the bacterial growth.
Without spoiling, we can just say that the previous model confirms the intuition we could have : while oxygen can't easily move into the medium, the growth on the surface area is sufficiently more important than in the medium to neglect the first one. More precisely, our bacteria is colorful and the surface will be entirely covered before internal bacteria have time to develop enough to be visible. Finaly, when we could hope to observe internal bacteria due to their color, the surface bacteria have already done an opaque layer.
So, this part aims at predicting over time the bacterial population growth on an ellipsoidal object - a fake lemon in practice -.
More precisely, we have to determine the initial proportion of bacteria threshold from which we are sure that the population will never extinct...if this threshold exists !
- An important aspect of our project is the transition from green to yellow, in order to emulate the lemon maturation.
To do this, we use a fusion protein. Before starting the fusion of the two -blue and yellow- chromoproteins, we have to check both compatibility. That's why we will then discuss the structural modeling of the proteins. We used here the bioinformatics tools developed by the Swiss Institute of Bioinformatics and by the Xu group of Chicago University.
We would have like modeling the salycilate inducible system to better understand this system but, unfortunatly, we do not have time anymore at last...
- Last but not least, the lemon scent !
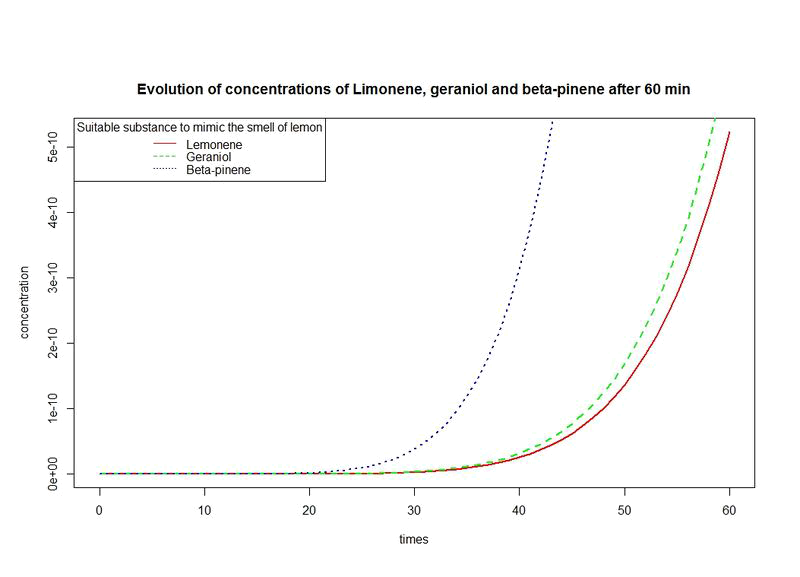 Actually, we have modified E.coli in order to let her produce pinene, limonene and Geraniol but thus do not ensure us that we will smell the lemon fragance.
Actually, we have modified E.coli in order to let her produce pinene, limonene and Geraniol but thus do not ensure us that we will smell the lemon fragance.
As De gustibus non est disputandum !, we need criteria to assess the scent of lemon; we proceed as follows:
- We evaluate the evolution of the concentrations of this three fragance over time.
- Give a procedure for estimating a suitable time interval in which we can say that we got the smell Citronel as Desired.
Note : We have tried to illustrate our work with figures which could be understood for themselves, without knowing the method to get it. Morever, we will just add one or two -for the warrior- star(s) to parts which need more mathematical background in order to don't disgust not-mathematical readers from mathematics.
 "
"




