Team:Oxford/progress2
From 2014.igem.org
Modelling Progress
Week 5 Day 5
Oliver says:
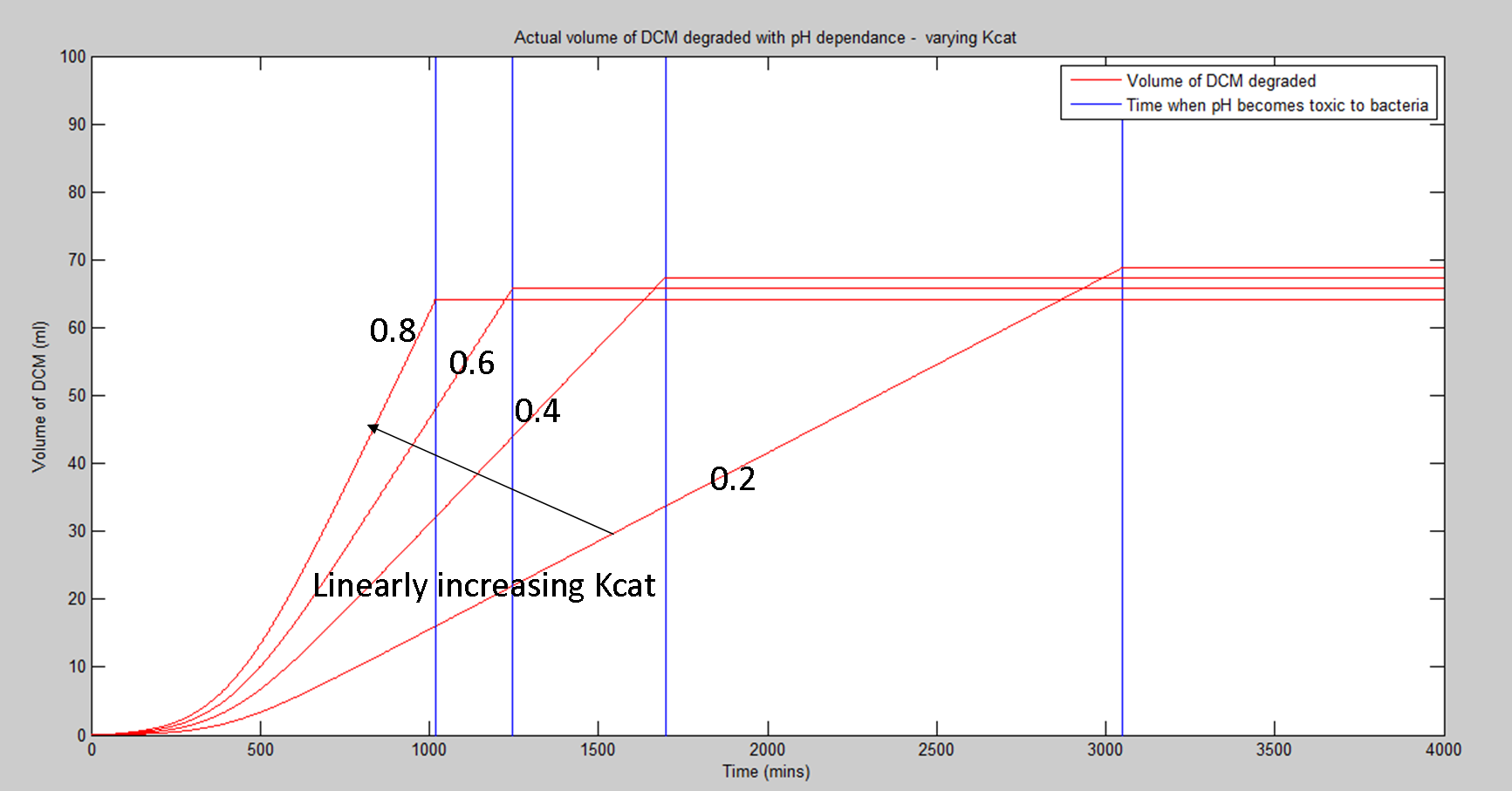
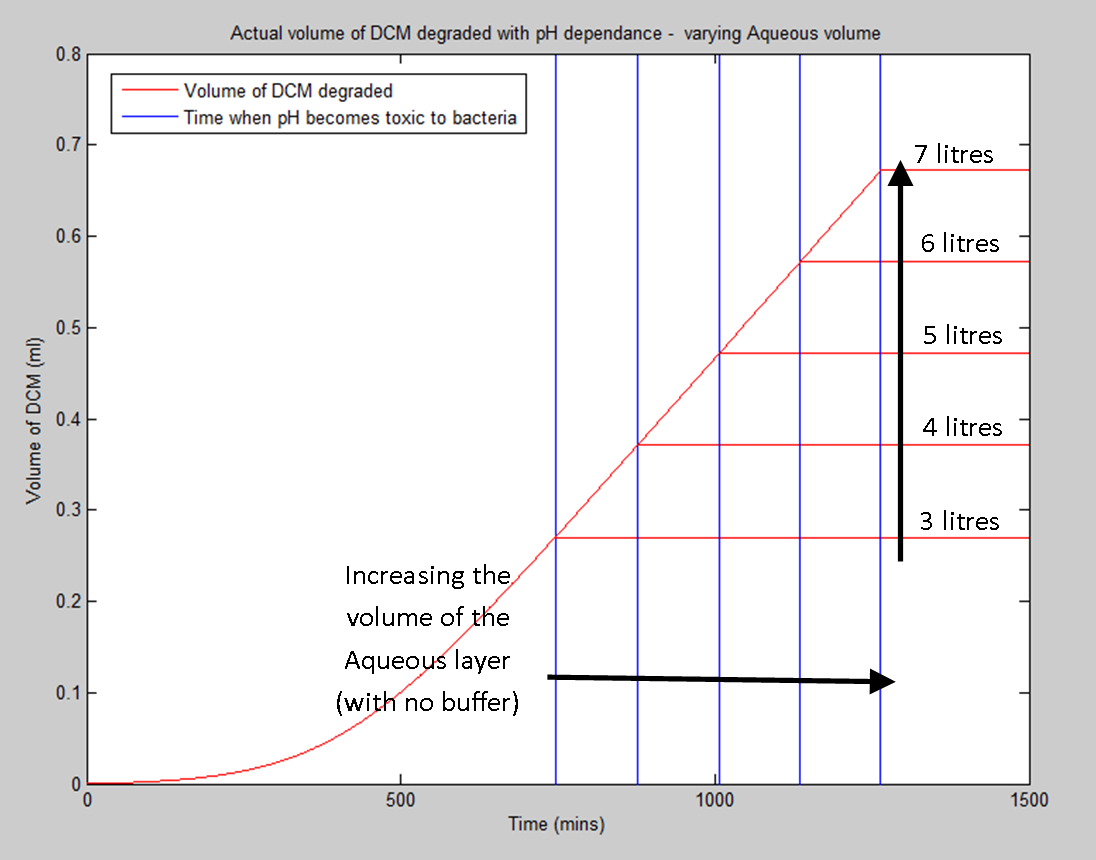
Today I will be working on adapting the result that I got yesterday (the pH dependent volume of DCM degraded) by varying the parameters that we are either uncertain of or can vary either mechanically (eg. amount of water added) or biologically (eg. Kcat through directed evolution) to analyse exactly how these parameters affect the overall important response of our system.
Leroy says:
Today and yesterday was spent further developing the compartment diffusion model. The model is now able to plot multiple species and highlight points of collision. The data which I hope to get out of this simulation is the ability to predict the interactions and accumulations of products during the metabolic pathway. The biochemists have told me, however, of the existence of a software package called GEPASI which will help me do this more effectively. Thus, my reaction model will likely only be used to illustrate the presence and likelihood of collisions between molecular species.
Week 5 Day 4
Oliver says:
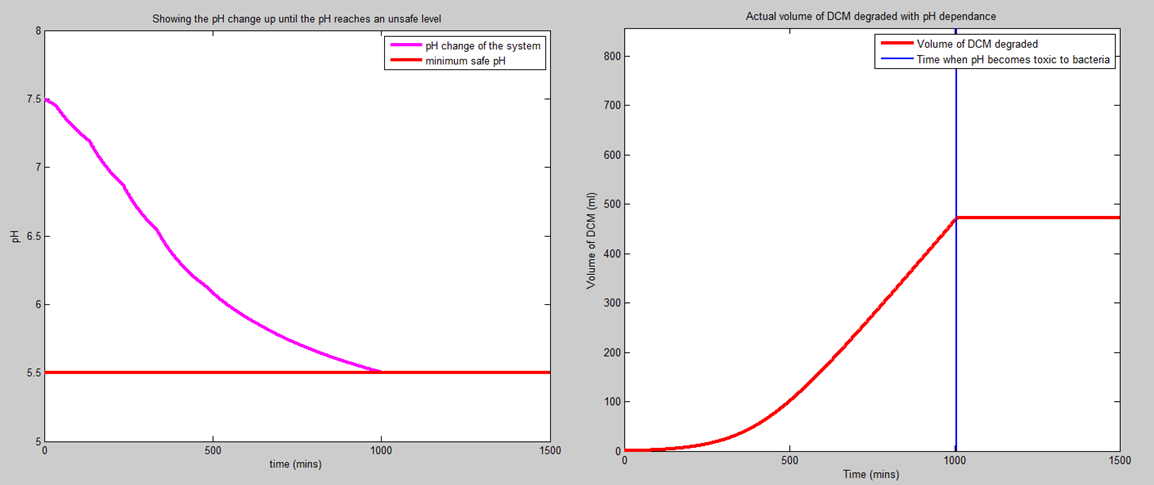
Today I have been working on the pH dependence of the system; namely making sure that the model doesn't assume that the bacteria will keep on degrading the bacteria once the pH reaches a level that the bacteria are unable to live in.
This idea will be extended to allow us to either calculate how much buffer to add to the aqueous layer or to allow us to calculate the amount of water to add to the aqueous layer so that in the lifetime of the bacteria, the pH doesn't drop to a level that would kill the bacteria prematurely.
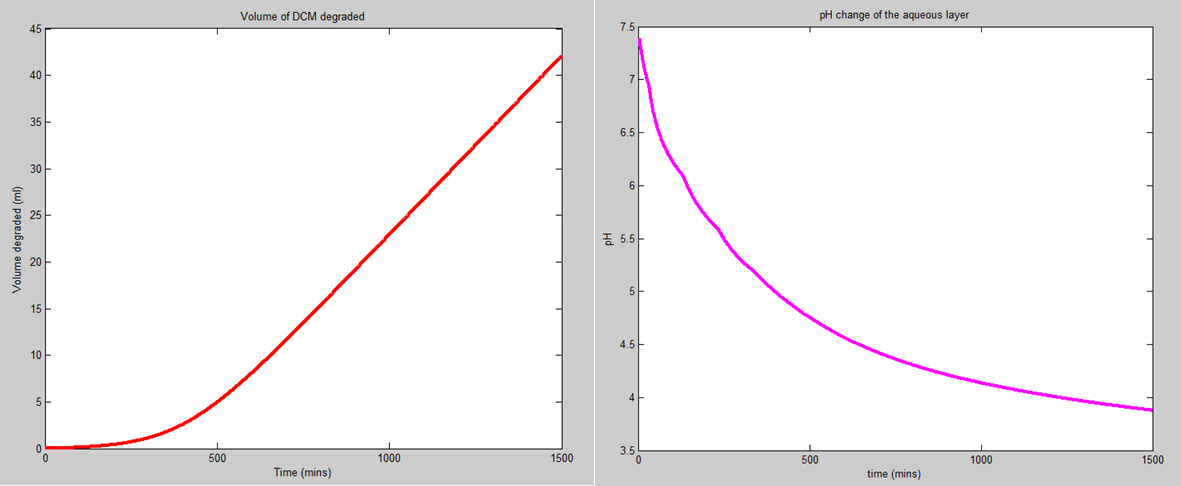
As you can see from the plots, the predictions for the amount of DCM that we should be able to degrade are quite promising. A major extension of this idea is that for our bacteria, Kcat is around 0.6 (molecules of DCM degraded per second by each dcmA molecule). This will be categorised more accurately by the biochems and their assays. However, for some degradation reactions in biology, the Kcat values are closer to 100000. As the volume of DCM degraded is directly proportional to the Kcat value, this hints that if we applied this system to breaking down other harmful substances with other bacteria, we could dramatically improve the rate at which we break down harmful substances, developing a new form of bio-remediation.
Week 5 Day 3
Oliver says:
Gave the presentation with Glen this morning to the Biochemistry Department. It was very good experience for giving presentations and for answering the questions that come afterwards.
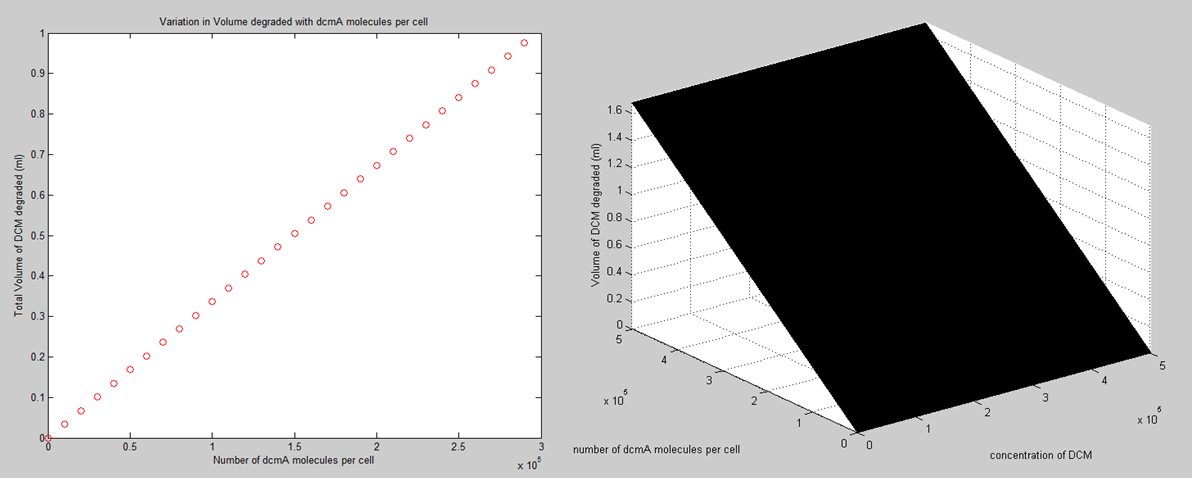
The afternoon was spent looking into the variations in how the total amount of DCM that we should be able to degrade is affected by the number of dcmA molecules in the cell. This is obviously a linear relationship that depends on the amount of time that you let the system run for. This is shown in a time varying 3D plot, this is shown on the right below.
This is an important consideration because when we are doing things like adding microcompartments to the cell, we need to analyse how this will affect the total percentage of cellular protein that will be dcmA and will therefore be utilized in breaking down the DCM.
The eventual plan is for the models that Matt and Leroy have been doing to coincide with mine. They should be able to increase the accuracy to which we know some of the terms in the equations.
Leroy says:
Today was spent further developing the compartment model of diffusion which I started yesterday. The model is now able to simulate any number of realizations of two different species of molecules with different properties. In addition to this, some of the molecules have been constrained to appear only in the microcompartment (DcmA) while some of the molecules are created anywhere within the space and can flow through the microcompartment unhindered.
Week 5 Day 2
Oliver says:
Spent the day on SolidWorks in the Engineering department. I built a 3D model of the product that we plan to build to allow us to visualize it and spot any flaws in the design. It also allows us to explain the project to potential users. It will be useful for the presentation to the Biochemistry Department tomorrow lunchtime.
Matt says:
Produced a new stochastic model which has a step input of C (DCM) after 50s which activates the production of B (DCMR), the equations were taken from Antonis's lecture notes about gene expression. I included a basal rate of B so that there was a small amount produced even when there is no C present.
Leroy says:
This was my first day back after taking about a week off for family reasons. Matt has continued the stochastic modelling which I started off on Friday of Week 3. I spent today developing the diffusive aspect of the stochastic modelling and have successfully modeled the diffusive motion, according to the Smoluchowski equations, of DCM and DCMA within the microcompartment. The model is such that DCMA molecules cannot diffuse through the microcompartment but DCM molecules can move in and out unhindered due to their small size.
Week 5 Day 1
Matt says:
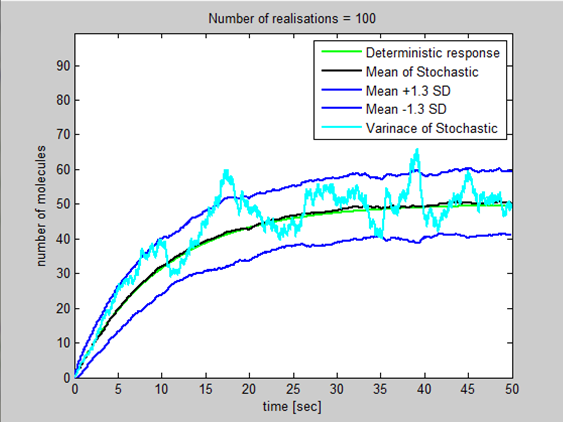
I went through the derivation of a general stochastic model today and proved that the steady state variance=steady state mean=k2/k1 where k2=production constant and k1=degradation constant. I introduced this variance into ProductionDegradation3 then using Standard deviation=(Variance)^0.5 introduced a basic upper/lower bound for the population by adding and taking away 1.3 standard deviation to the mean. This gives us a confidence interval of 90% that the population will lie between the 2 blue lines. (Assuming the variation in population around the mean can be modelled using the normal distribution)
Oliver says:
Corrected the model that I built on Wednesday last week. That model was concerned with intracellular HCl concentration and therefore intracellular pH. However, after speaking to Ciaran, we think it's more realistic to assume that the cells have a chloride efflux pump. Therefore, we should be concerned with both the HCl concentration and pH outside of the cell. This obviously also depends on the volume of the aqueous layer, so the pH change that I've calculated won't be the real pH change of the system due to the fact that we will add a buffer solution to the container. The new volume of DCM degraded and pH change (without buffer) curves are shown below. The amount of DCM that we can degrade will be proportional to the number of happy bacteria that we have in the system and the length of time that we can keep them alive for.
 "
"