Before we get started:
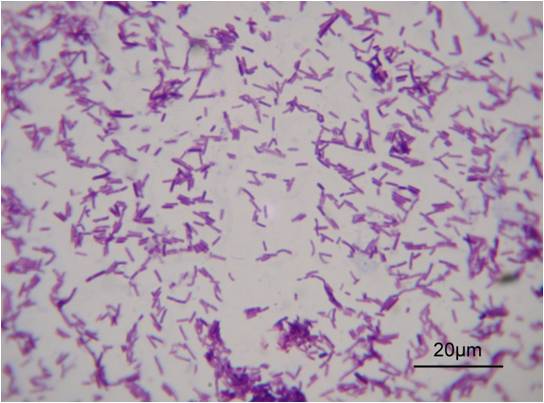
The Gram positive L. casei., LB23
The purpose of our study attempts to construct an E.coli-Lactobacillus shuttle vector plasmid with an expression cassette consisting of a strong promoter, a signal sequence, and a heterologous model protein gene. We put the plasmid inE. coli, our testing bacteria to conform the feasibility of our study and finally the plasmid will be inserted into L. casei in "HOPE".
Generally, lactic acid bacteria (LAB) are regarded as safe and useful commensal bacteria, which are known as probiotics and starters for food fermentation. In recent years there has been an upsurge of interest in the genetic manipulation of lactic acid bacteria (LAB).
Many studies on Lactobacillus casei subsp. have been carried out using the plasmid-free strain BL 23, ATCC 393 (pLZ15-)[4]. BL 23 and its relatives have been used as a host for genetic modification and applied for live delivery agents. For example, the single chain variable fragment of an antibody that binds to a major adhesion molecule of Streptococcus mutants was expressed on the cell surface and showed protective effects against colonization with the pathogen [5]. Thus, it is clear that the development of a highly efficient protein-secreting system in this strain would be important and beneficial.
Moreover, L. casei might be a better live delivery agent because the optimal growth temperature of L. casei is the same as the body temperature of mammals. Thus, BL 23 is no doubt a suitable bacteria host for our food-grade protein secreting system in “HOPE”.
So how did we do it?
Bacterial Strains and Growth Conditions
L. casei BL23 and recombinant strains were grown in Mann-Rogosa-Sharp (MRS) medium at 37℃.
Erythromycin (5 μg/ml) was added to MRS for the selection of recombinant L. casei strains. For pH control, 50 mM carbonate buffer was supplemented into the medium at certain ratios (NaHCO3:Na2CO3). The commonly used cloning host of E. coli DH5α(TaKaRa, Shiga, Japan) was grown in LB with or without 100 μg/ml ampicillin. L casei, BL 23 were maintained in a 10% glycerol stock collection at -80 °C and were subcultured in MRS medium (Oxoid) when required.
Bacterial strains used in this study:
Strain code | Strain | Source BL 23 | L. casei ATCC 393 (pLZ15-) | [Y.C Tsai] NYMU, Taiwan
Construction of Plasmids and Transformation of L. casei
The sequences of all PCR primers used in this study are listed in the following Table 2. PCR products and plasmids were digested with restriction endonucleases followed by ligation and transformation to E.coli DH5α. In the cloning of E.coli DH5α we used a high copy number plasmid named E. coli, pUC19 After harvesting L. casei plasmid by digesting pUC19, we use a E-coli-Lactobacillus shuttle vector, pLP402, to perform our desired features. And the transfer process was carried out by electrophoresis.
PCR primers used in this study:
Primers Sequences Restriction sites IGM289 cccaagcttagatctgattacaaaggctttaagcagg HindIII, BglII IGM290 gggctcgaggcccggttgttcgcggccgcttttgttaagaattttatttcataacattagcgg XhoI, NotI IGM291 cccgtcgacgcccggttgttcgcggccgcttcggttatactattcttgcttgata SalI, NotI IGM292 ggggaattcctgcagggatccaaacttgattgcataatctttcttcc BamHI, PstI, EcoRI IGM350 acatattttatgtttggagggtattggatg IGM351 catccaataccctccaaacataaaatatgt IGM468 ccccggatccgagcaggggccagtacagccg BamHI IGM478 ccccctcgagttagcttttcattttgatcatcatgta XhoI IGM479 gcgaaatccaagcaaaggcgagcaggggccagtacagccg IGM480 cggctgtactggcccctgctcgcctttgcttggatttcgc IGM482 cggctgtactggcccctgctcggatccgagtttgtgtccgcctttgcttggatttcgc BamHI
Expected results~
Construction-flow diagram of expression vectors. MCS: multiple cloning site; Rep: replication protein; Emr: erythromycin resistance; Ampr: ampicillin resistance

Gene map of plasmid pMC11 and putative replicons, ORFs.RepA_1 and RepB_1 are replication initiation proteins; RepA and RepB are putative replication proteins. For trehalose operon, PG-R is trehalose operon repressor, TreC is trehalose-6-phosphate hydrolase and TreB-PTS is PTS system trehalose-specific IIBCA components.
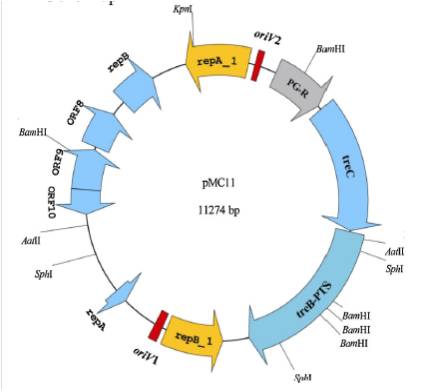
C, D Replication origins fragment of pMC11. Repeats of oriV1 (iterons contain 4.5 direct repeats) and oriV2 (iterons contain 3.5 direct repeats) and invert repeat are diagrammed. Promoter elements and ribosome binding sites (RBS) of repA_1 and repB_1 genes are indicated.

 "
"




















