Before we get started:
Biofilm formation is part of the reason S. mutans is so devastating to oral health (see [Project Overview] for more information). For adequate oral protection, biofilm formation is a problem we must solve, and we can do so externally, via our probiotic (see [Cleanse: Antibiofilm]), or internally, by altering the genome of S. mutans. This means we need a way to silence the appropriate proteins.
So how are proteins silenced? As we know, proteins are translated from single-stranded mRNAs. Just like eukaryotic cells, bacteria can selectively silence mRNA by transcribing sequences of sRNA, or bacterial small RNA.
These small RNA molecules are usually less than 50bp long, and function by binding to the specified, complementary mRNA strand to prevent ribosome attachment. We decided that targeted sRNA would give us a good shot at silencing proteins in S. mutans.
Next up, we need to find some proteins to silence!
So how did we do it?
After careful literature search, we found two candidates:
Histidine kinase 11 (HK11) is a sensor kinase from a two-component signal transduction system. Literature indicates that a knock-out of HK11 resulted in dramatically decreased biofilm formation.
G protein in S. mutans (SGP) is involved in the regulation of intracellular GTP/GDP ratio, response to stress, and other diverse cellular functions. As with HK11, a knock-out of SGP resulted in dramatically decreased biofilm density.
In order to silence these two proteins with sRNA, we synthesized a 24bp strand of non-coding DNA, and transcribed it into the corresponding RNA. This small RNA sequence is our artificial sRNA, and will bind to the TIR (translation initiation region) of our target mRNAs. This prevents the target mRNA from undergoing translation.
The final thing we need is something to help smooth out the whole process. The MicC scaffold is a specially constructed sequence which recruits the Hfq protein. The Hfq protein then helps our sRNA hybridize with its target mRNA, while stabilizing the sRNA-mRNA complex. Note that, since we want the products in RNA form, we also do not need an RBS (ribosome binding site).
At last, our circuit is complete!
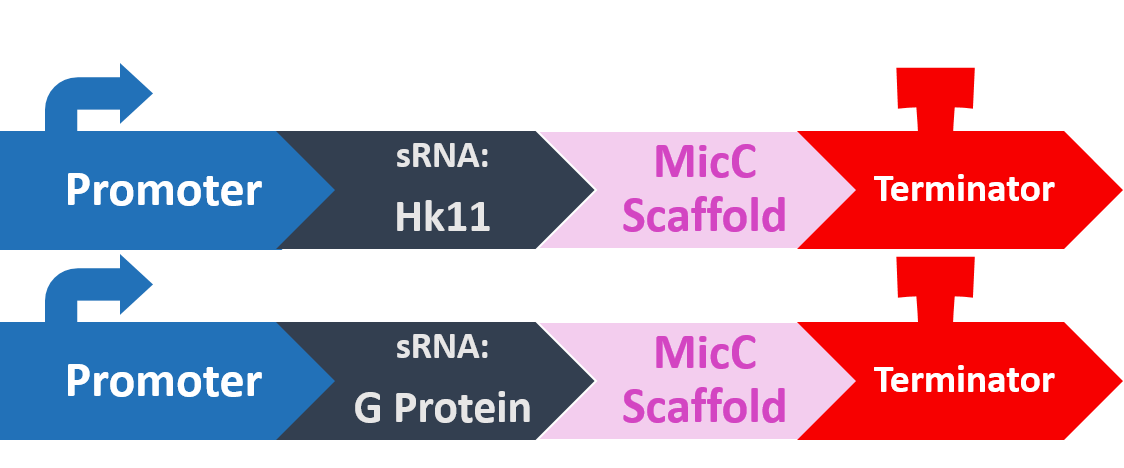
Here is what our product would look like:

As mentioned, the sRNA part would bind to the TIR of the target mRNA, while the MicC scaffold recruits the Hfq protein to stabilize the entire structure.
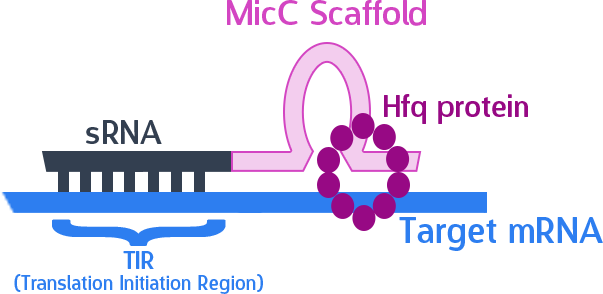
Putting it to the test!
Having a circuit is only the start! Now we must test how well our circuits function. For the sRNA protein-inhibition module, a crucial test is of course how well the system performs, which is measured by the biofilm formation before and after transformation.
Does sRNA silencing work?
To test our sRNA function, we first cultured S. mutans so they have a chance to form a layer of biofilm. After biofilm is formed, we discarded the supernatant, and stained with a histological stain called Crystal Violet (CV). CV was used primarily to distinguish between Gram bacteria types, and have a strong affinity to biofilms, whose composition contains similarities with cell walls. After drying the sample and removing excess stain, we dissolved the biofilm and measured OD, to determine the amount of biofilm that has formed.
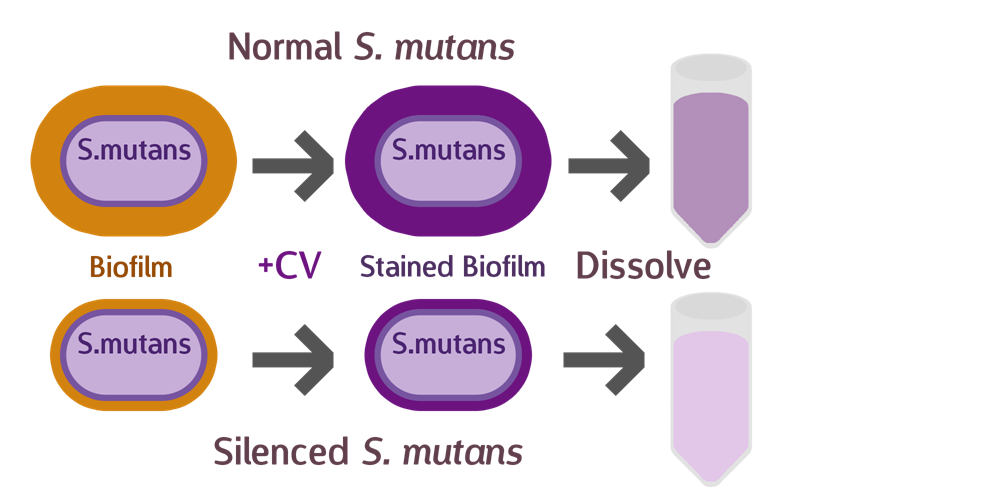
Theoretically, if our sRNA silencing works, we should see less biofilm (and less CV) when compared to the wild type.
Here is the details of our method:
- Incubated a liquid culture of S. mutans overnight.
- Incubated a fixed quantity of S. mutans at 37°C in 96-well polystyrene plate for 24hrs.
- Discarded the supernatant and rinsed the wells once with sterile distilled water.
- Stained S. mutans with crystal violet.
- Washed wells once with ddH2O to remove excess dye and air dried for 3hr.
- Biofilm dissolved by 95% ethanol.
- Based on the hypothesis that S. mutans quantity is proportional to the quantity of biofilm, we will measure the absorbance of biofilm molecules at OD 575nm and compare to the wild type S. mutans.
- According to the result, we can see if the biofilm formation has decreased.
 "
"




















