Team:NYMU-Taipei/notebook/labnotes
From 2014.igem.org
(Difference between revisions)
m |
|||
| Line 97: | Line 97: | ||
<h2>Parts from iGEM took kit</h2> | <h2>Parts from iGEM took kit</h2> | ||
<h3>Out line</h3> | <h3>Out line</h3> | ||
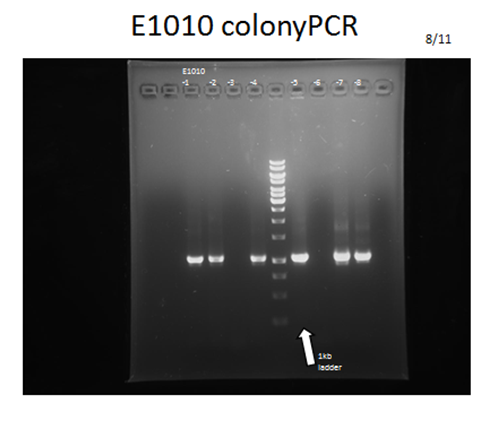
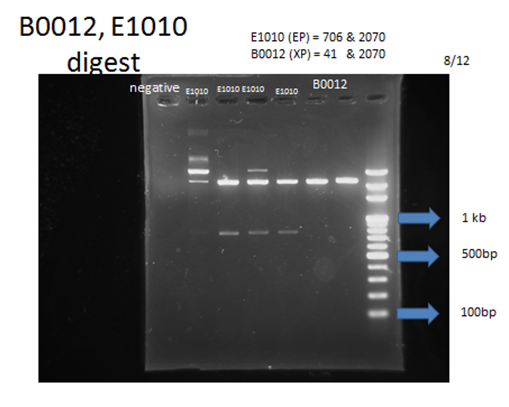
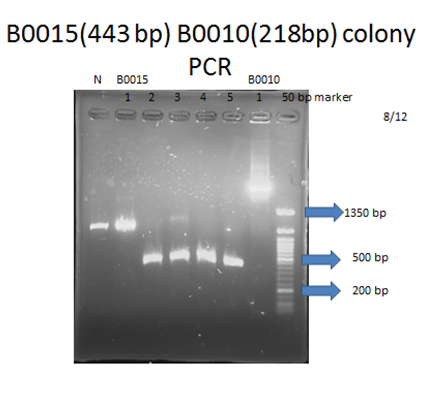
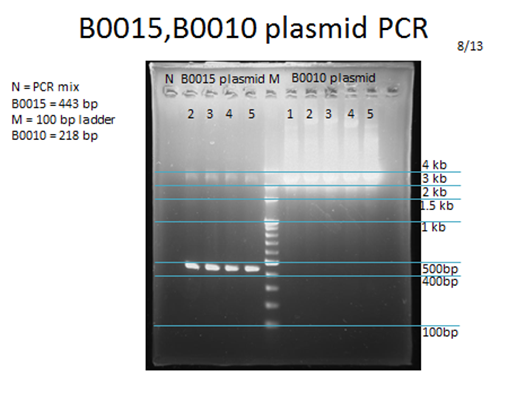
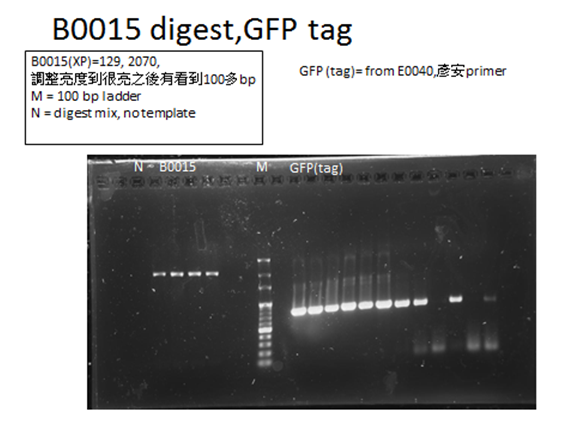
| - | <p>For each part to be used in testing circuit:<br>BBa_J23100 , BBa_E1010, BBa_B0015 | + | <p>For each part to be used in testing circuit:<br>BBa_J23100 , BBa_E1010, BBa_B0015</p> |
| - | </p> | + | |
<img src=''><p>circuit 圖</p> | <img src=''><p>circuit 圖</p> | ||
<p>After transforming these plasmids into our competent cell, DH5a, we would take 3 steps to examine if we got the right product:</p> | <p>After transforming these plasmids into our competent cell, DH5a, we would take 3 steps to examine if we got the right product:</p> | ||
| Line 118: | Line 117: | ||
<img src='/wiki/images/c/cd/NYMU14_note_4c_r4.PNG' style='width:450px;'> | <img src='/wiki/images/c/cd/NYMU14_note_4c_r4.PNG' style='width:450px;'> | ||
<img src='/wiki/images/e/eb/NYMU14_note_4c_r5.PNG' style='width:500px;'> | <img src='/wiki/images/e/eb/NYMU14_note_4c_r5.PNG' style='width:500px;'> | ||
| + | <h2>Parts that needed extra PCR</hr> | ||
| + | <h3>Outline</h3> | ||
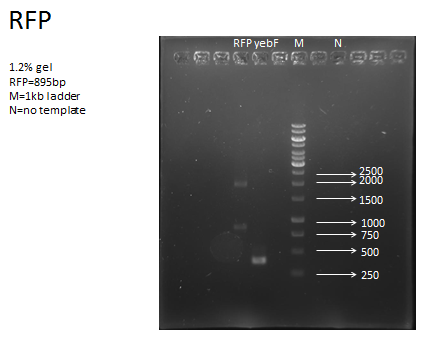
| + | <p>Two parts needed extra primer in order to get them:<br>yebF, RFP, and M102 orf19</p> | ||
| + | <img src=''><p>circuit 圖</p> | ||
| + | <p>And for each part, we do two kinds of PCR , one using Dream Tag, only to check the length to ensure we got our part. And then we used KOD enzyme, together with magnesium cation, to check the length, furthermore, do gel extraction to get the part. That’s because KOD enzyme leads a more accurate PCR.</p> | ||
| + | <ol> | ||
| + | <li>For yebF, we designed primers to get the sequence we want yebF (its own ribosomal binding site included) from Escherichia coli MG1655.<br>Front Primer : tttctagagGGAGAAAAACATGAAAAAAAGAGGGGCGTT(with Xbal cutting site)<br>Reverse Primer : attctgcagcggccgctactagtACGCCGCTGATATTCCGCCA(with Spel and Pst1 cutting site)<br>The PCR product should be 389bp.</li> | ||
| + | <li>Because the need of recombinant protein, the RFP downstream of yebF need to be specially designed in order to leave scar with multiple of 3. E1010 was used as template, and primers with specially designed cutting site is made:<br>Front Primer: tttctagaGCTTCCTCCTCCGAAGACGTTATC<br>And normal iGEM reverse primer, VR</li> | ||
| + | </ol> | ||
| + | <h3>Process</h3> | ||
| + | <p>While doing RFP PCR, we got extra band.<br>By adjusting PCR program, we finally got correct band.</p> | ||
| + | <img src='/wiki/images/6/6b/NYMU14_note_4c_r6.PNG' style='width:450px;'> | ||
| + | <h3>Result</h3> | ||
| + | <img src='/wiki/images/1/12/NYMU14_note_4c_r7.PNG' style='width:450px;'> | ||
| + | <img src='/wiki/images/f/fc/NYMU14_note_4c_r8.PNG' style='width:450px;'> | ||
| + | <img src='/wiki/images/6/63/NYMU14_note_4c_r9.PNG' style='width:450px;'> | ||
</div> | </div> | ||
<!-----------------------------------------------------------> | <!-----------------------------------------------------------> | ||
Revision as of 12:50, 29 September 2014
X
Next ⇒
⇐ Prev
...
introduction
control
communication
cohesion
completion
function test
introduction :P introduction :P introduction :P introduction :P
introduction of labnote
樹狀圖 timeline 順序圖
under construction
under construction
under construction
under construction
under construction
under construction
under construction
under construction
under construction
under construction
under construction
under construction
under construction
 "
"