Team:NCTU Formosa/project
From 2014.igem.org
(→Overview) |
|||
| Line 25: | Line 25: | ||
====Pesticide hazards==== | ====Pesticide hazards==== | ||
| - | |||
====Solution to both questions==== | ====Solution to both questions==== | ||
| Line 33: | Line 32: | ||
| - | |||
===PBAN=== | ===PBAN=== | ||
Revision as of 17:05, 29 August 2014
Contents |
Overview
Impact of pest
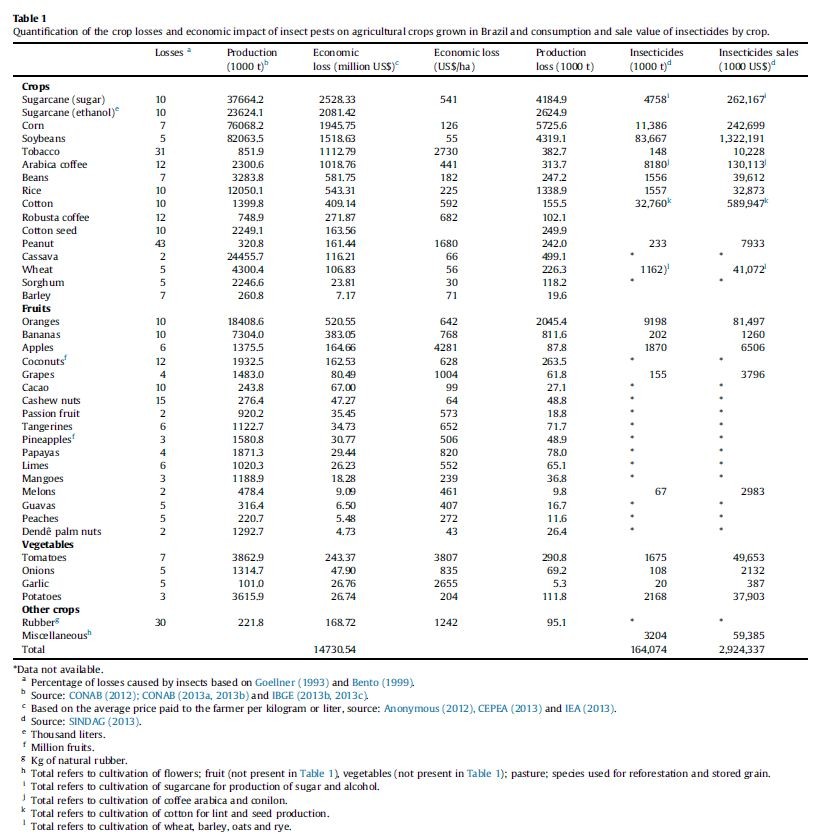
In agriculture, insect problem has existed for a long time, and has been difficult to be solved, all of the agricultural products all over the world every year cause serious harm and damage caused by these insects bulky and difficult to estimate.
Losses caused by pests on crops included direct damage, drug costs and the labor costs, The most difficult to estimate the loss of pesticides on human health hazards caused by consuming a lot more medical resources.
We can see this a credible chart, insect damage recorded in Brazil region, pests generally reduce crop yields by 10 percent, which is each crop have reduced the value of hundreds of millions. Our goal is committed to reducing agricultural losses, safeguard human health, and to maintain ecological balance.
Pesticide hazards
Solution to both questions
In order to solve the pest need to use pesticides, but the use of pesticides will destroy the Earth and cause harm to humans. Thus there has been conflicting issues, and has been difficult to be solved. In order to solve the problems we mentioned, we came up with a practical, inexpensive way to solve. Figure 1 is a diagram of our device, after which we will mention how this device to bring better results for our plan and improve his efficiency.
PBAN
Introduction
content
content
content
content
Design of Red promoter
content
Reference
- part BBa_I15008;MIT Registry of Standard Biological Parts
- part BBa_I15009;MIT Registry of Standard Biological Parts
- Levskaya, A. et al .(2005). Engineering Escherichia coli to see light. Nature, 438(7067), 442.
- Kehoe DM, Grossman AR (1996) Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science 273(5280):1409–1412
- Yeh KC, Wu SH, Murphy JT, Lagarias JC (1997) A cyanobacterial phytochrome two-component light sensory system. cience 277 (5331):1505–1508
- Dutta R, Qin L, Inouye M (1999) Histidine kinases: diversity of domain organization. Mol Microbiol 34(4):633–640
- Forst SA, Roberts DL (1994) Signal transduction by the EnvZ–OmpR phosphotransfer system in bacteria. Res Microbiol 45(5–6):363–373
- Thomas Drepper, Ulrich Krauss,Sonja Meyer zu Berstenhorst, Jörg Pietruszka, Karl-Erich Jaeger.(2011).Lights on and action! Controlling microbial gene expression by light. Appl Microbiol Biotechnol, 90:23–40 DOI:10.1007/s00253-011-3141-6
Biobrick Design
Introduction
Mechanism
Design
Reference
- Torsten Waldminghaus, Nadja Heidrich, Sabine Brantl and Franz Narberhaus .(2007). FourU: a novel type of RNA thermometer in Salmonella . Molecular Microbiology , 65(2): 413–424 DOI:10.1111/j.1365-2958.2007.05794.x
- part BBa_K115002;TUDelft Registry of Standard Biological Parts
Device
Introduction
Mechanism
Design
Reference
- Xu, S.; Montgomery, M.; Kostas, S.; Driver, S.; Mello, C. (1998). "Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans". Nature 391 (6669): 806–811 DOI:10.1038/35888
- Jörg Vogel , Ben F. Luisi.(2011). Hfq and its constellation of RNA. Nature Reviews Microbiology, 9:578-589
- E.K. Jocelyn, S.G. Elliott , T.K. Stephen, "Lewin's Genes X.-10th ed.", Jones & Bartlett, Sudbury, MA, 2011.
- Karen M. Wassarman.(2002). "Small RNAs in Bacteria: Diverse Regulators of Gene Expression in Response to Environmental Changes". Cell, 109:141–144
- Hongmarn Park, Geunu Bak, Sun Chang Kim & Younghoon Lee.(2013). "Exploring sRNA-mediated gene silencing mechanisms using artificial small RNAs derived from a natural RNA scaffold in Escherichia coli ". Nucleic Acids Research,Vol. 41, No. 6, 3787-3804 DOI:10.1093
- Vandana Sharma, Asami Yamamura & Yohei Yokobayashi.(2011). "Engineering Artificial Small RNAs for Conditional Gene Silencing in E. coli". ACS Synthetic Biology
Cover image credit: DVQ
 "
"