Team:Michigan Software/Project
From 2014.igem.org
(→Methods) |
|||
| Line 52: | Line 52: | ||
• is built on a well-documented , easy to learn framework. | • is built on a well-documented , easy to learn framework. | ||
| - | Our web application can be downloaded from Github and easily installed on any computer with Django/Python. Our implementation uses an embedded SQLite database, although a number of databases can be used. | + | Our web application can be downloaded from Github and easily installed on any computer with Django/Python. Our implementation uses an embedded SQLite database, although a number of databases can be used. |
[[File:concept_map.png]] | [[File:concept_map.png]] | ||
| Line 62: | Line 62: | ||
databasemodels\<br> | databasemodels\<br> | ||
templates\<br> | templates\<br> | ||
| + | <br><br> | ||
| - | + | In order to create the database for first-time use, check models.py in the "databasemodels" folder. If you make alterations to this file, or just want run our implementation, navigate to Michigan14-branch1\ProtoDB in your command-line terminal and type "python manage.py syncdb". This will create/update the SQLite database. | |
| - | + | ||
| - | Michigan14-branch1\ | + | ProtoDB (the second one) contains the settings files and the URL file. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==Success== | ==Success== | ||
Revision as of 23:34, 17 October 2014
Contents |
Description
Choosing reliable protocols for new experiments is a problem laboratories routinely face. Experimental practices differ immensely across laboratories, and precise details of these practices may be lost or forgotten as skilled members leave the lab. Such fragmentation in protocol methods and their documentation often hampers scientific progress. Indeed, there are few well-defined protocols that are generally agreed upon by the scientific community, in part due to the lack of a system that measures a protocol’s success. In turn, the lack of commonly accepted protocols and inadequate documentation affects experimental reproducibility through method inconsistencies across laboratories. Review studies even estimate that only 10-25% of published scientific results are reproducible, and up to 54% of materials/antibodies/organisms are not identifiable for reproducibility studies (https://peerj.com/articles/148/). This is an alarming figure and suggests that many of the results we base continued research and development on are inaccurate or cannot be replicated independently.

 To attempt to address these problems, we set out to build a database that integrates a crowd-sourced ratings and comments system to clearly document, rate, elaborate, review, and organize variants of experimental protocols in order to increase the likelihood of reproducible scientific results. This would streamline some of the confusion associated with finding and replicating protocols by creating a database where investigators can upload protocols, comment on them, and even use a rating system to allow the best protocols to rise to the top. Before starting, we designed a survey (check it out here) to poll a range of scientific researchers on their experiences trying new protocols. We disseminated it across our networks and over social media to judge the interest and usefulness of our project. We found that among a diverse and experienced set of respondents, every single scientist has struggled with replicating protocols from other experimenters, with >50% of respondents having difficulty more than 25% of the time.
To attempt to address these problems, we set out to build a database that integrates a crowd-sourced ratings and comments system to clearly document, rate, elaborate, review, and organize variants of experimental protocols in order to increase the likelihood of reproducible scientific results. This would streamline some of the confusion associated with finding and replicating protocols by creating a database where investigators can upload protocols, comment on them, and even use a rating system to allow the best protocols to rise to the top. Before starting, we designed a survey (check it out here) to poll a range of scientific researchers on their experiences trying new protocols. We disseminated it across our networks and over social media to judge the interest and usefulness of our project. We found that among a diverse and experienced set of respondents, every single scientist has struggled with replicating protocols from other experimenters, with >50% of respondents having difficulty more than 25% of the time.
 Additionally, our survey identified that unclear language and missing steps of protocols as the greatest contributors to the irreproducibility of protocols. Furthermore, 100% of respondents indicated they would use a database like this to browse and download protocols, and over 85% indicated they would upload and maintain their own protocols if such a site existed. With these data and interest in hand, we set out to build ProtoCat.
Additionally, our survey identified that unclear language and missing steps of protocols as the greatest contributors to the irreproducibility of protocols. Furthermore, 100% of respondents indicated they would use a database like this to browse and download protocols, and over 85% indicated they would upload and maintain their own protocols if such a site existed. With these data and interest in hand, we set out to build ProtoCat.
Aims
The aims of this project were threefold:
- To start an iGEM software team at the University of Michigan and populate it with a diverse population of students.
- To construct useful software for laboratory scientists. We designed the protocol database project based on our own and other's frustrations in identifying reliable and effective scientific protocols.
- To use our basic project idea to survey a wide range of scientific students and professionals to determine exactly what the best iteration of this project would do and adapt our goals as such.
Methods
Our project is based on Django, a free, open-source web framework, written in Python. Django allows us to create a simple, database-driven website which:
• can be hosted on any server (or locally).
• is modular and can be molded to specific individual needs.
• is built on a well-documented , easy to learn framework.
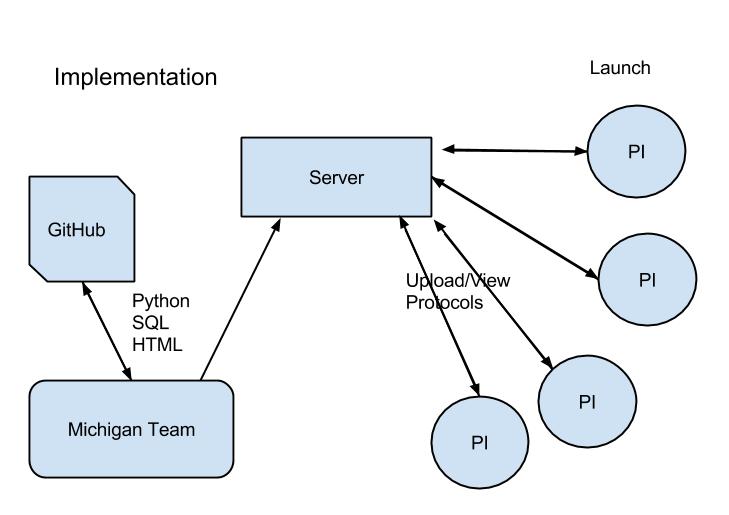
Our web application can be downloaded from Github and easily installed on any computer with Django/Python. Our implementation uses an embedded SQLite database, although a number of databases can be used.
Unzipping the folder will result in the following file structure:
Michigan14-branch1\
ProtoDB\
ProtoDB\
databasemodels\
templates\
In order to create the database for first-time use, check models.py in the "databasemodels" folder. If you make alterations to this file, or just want run our implementation, navigate to Michigan14-branch1\ProtoDB in your command-line terminal and type "python manage.py syncdb". This will create/update the SQLite database.
ProtoDB (the second one) contains the settings files and the URL file.
Success
- Formed a brand new iGEM team!
- Reached out to and engaged the scientific community to create awareness for the problem and prioritize the development of a solution.
- Created ProtoCat, a functional registry for laboratory procedures.
Future Directions
- variable protocols with integrated calculators
- vendor information
- in-app purchasing
- protocol timer
 "
"