Team:Heidelberg/pages/Parts
From 2014.igem.org
| Line 137: | Line 137: | ||
| Gp41-1||fastes known reaction ||88||38||t1/2=20-30s, 37°C, k=~1.8x10^-1 (s^-1); activity range 0 to 60°C||natural split intein, Cyanophage||[[#References|[7]]] [[#References|[8]]] | | Gp41-1||fastes known reaction ||88||38||t1/2=20-30s, 37°C, k=~1.8x10^-1 (s^-1); activity range 0 to 60°C||natural split intein, Cyanophage||[[#References|[7]]] [[#References|[8]]] | ||
|- | |- | ||
| - | |||
|} | |} | ||
Revision as of 21:14, 15 October 2014
Favorite Parts.
The iGEM Team Heidelberg 2014 had built a new biological system for the iGEM community integrating split-inteins. Intein splicing is a natural process that excises one part of a protein and leaves the remaining parts irreversibly attached. This great function allows you to modify your protein in numerous ways.
Creating a toolbox including all great functions and possibilities of inteins, we need a new standard for the scientific world of iGEM. This standard, the RFC of the iGEM Team Heidelberg 2014, allows us to easily and modulary work with split inteins.
Our favorite Parts represent the basic constructs of our toolbox – the Assembly and the Circularization construct, which are both tested in many methods and applications.
In the following we present you BBa_K1362000, the construct for circularization, BBa_K1362100 and BBa_K1362101, the N- and the C-construct for assembly. Take a look and visit the Partsregistry to read the associated documentation.
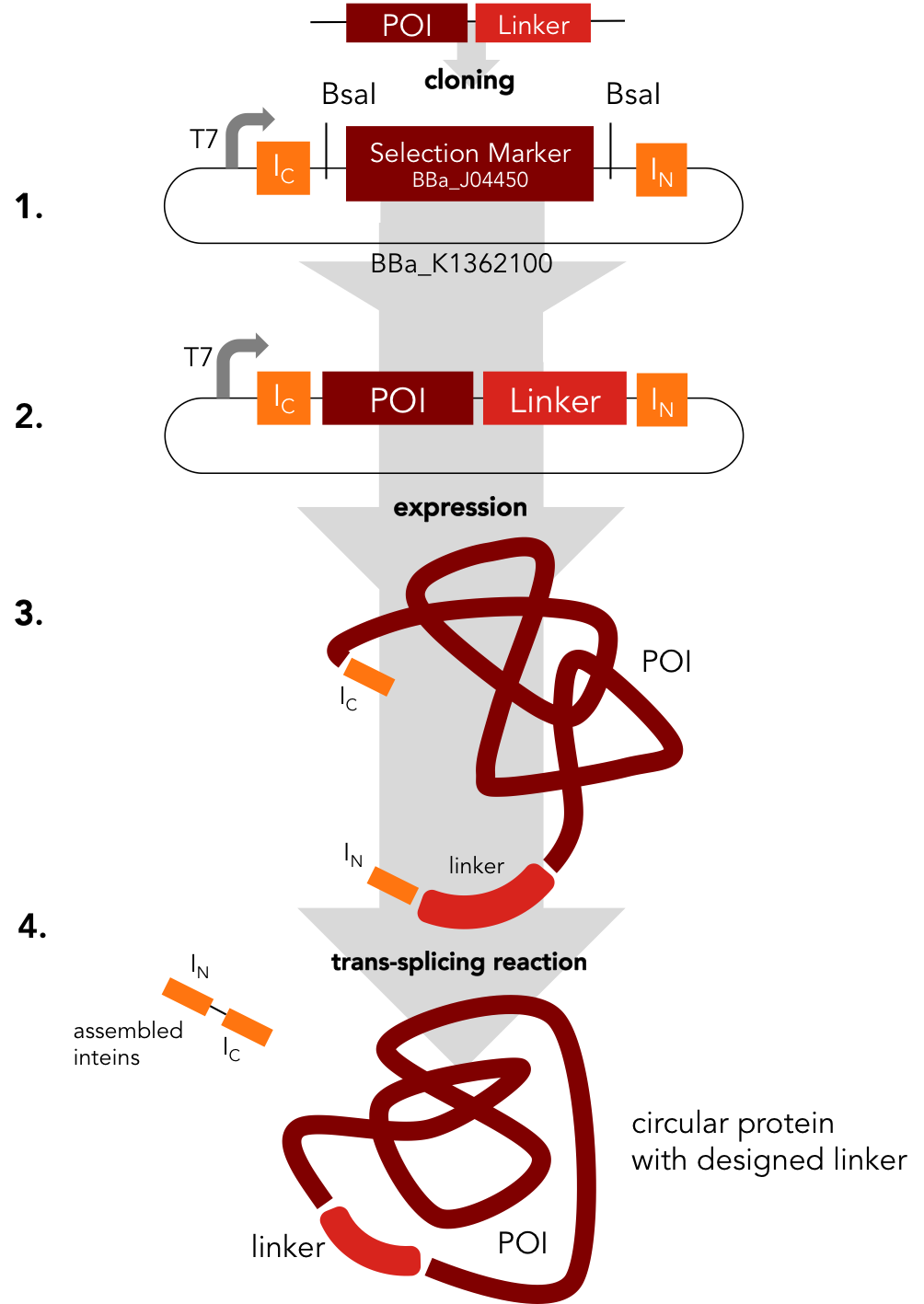
Circularization Construct. BBa_K1362000
BBa_K1362000
Placeholder
Assembly Constructs. BBa_K1362100 and BBa_K1362101
BBa_K1362100
This intein assembly construct is part of our strategy for cloning with split inteins. Inteins are naturally occuring peptide sequences that splice out of a precursor protein and attach the remaining ends together to form a new protein. When splitting those intein sequence into an N-terminal and a C-terminal split intein one is left with a powerful tool to post-translationally modify whole proteins on the amino-acid sequence level. This construct was designed to express any protein of interest fused to the Nostoc punctiforme DnaE N-terminal split intein.

BBa_K1362101
BBa_K1362101 is the corresponding C-terminal construct to BBa_K1362100. Upon coexpression or mixture of the N- and C-constructs protein splicing takes place and the N- and C-terminal proteins of interest are irreversibly assembled via a newly formed peptide bond.This mechanism can be applied for a variety of different uses such as the activation of a protein through reconstitution of individually expressed split halves. See our split sfGFP experiment and the respective parts in the registry for more information. Protein splicing offers many new possibilities and we hope to have set a foundation that you guys can build on!


Sample Data Page for our favorite Parts.
Circularization Construct. BBa_K1362000
Assembly Construct. BBa_K1362100 and BBa_K1362101
Placeholder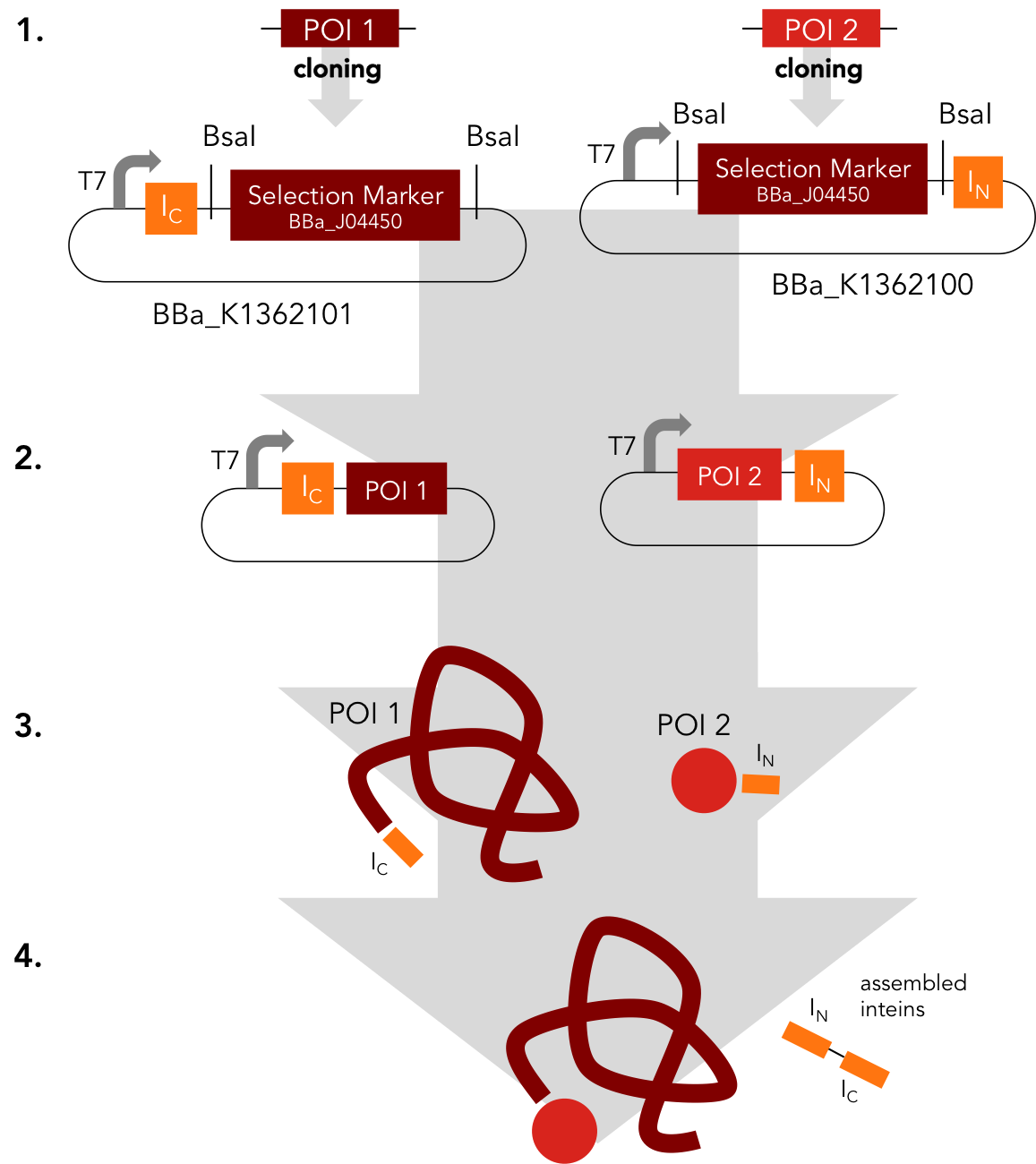

Intein libary
{{| class="table table-hover" |- !Split intein!!Special features!!!Nint!!!!Cint!!!!!Reaction propterties!!!!!!Origin!!!!!!!References |- | Npu DnaE||fast; robust at high temperature range and high-yielding trans-splicing activity, well characterised requirements||102||36||t1/2 = 63s , 37°C , k=~1x10^-2 (s^-1); activity range 6 to 37°C||S1 natural split intein, Nostoc punctiforme||[1] [2] |- | Ssp DnaX||cross-reactivity with other N-inteins, transsplicing in vivo and in vitro, high yields||||||k=~1.7x10^-4(s^-1); efficiency 96%||engineered from Synechocystis species||[3] [4] |- | Ssp GyrB|| very short Nint facilitates trans-splicing of synthetic peptides||6||150||k=~1x10^-4(s^-1), efficiency 40-80%||S11 split intein enginered from Synechocystis species, strain PCC6803||[4] [5] |- | Ter DnaE3||trans-splicing activity with high yields||102||36||k=~2x10^-4(s^-1), efficiency 87%||natural split intein, Trichodesmium erythraeum||[4] [6] |- | Ssp DnaB||relatively fast||||||t1/2=12min, 25°C, k=~1x10^-3(s^-1)||engineered from Synechocystis species, strain PCC6803||[2] |- | Gp41-1||fastes known reaction ||88||38||t1/2=20-30s, 37°C, k=~1.8x10^-1 (s^-1); activity range 0 to 60°C||natural split intein, Cyanophage||[7] [8] |- |}
 "
"