Team:Heidelberg
From 2014.igem.org
(Difference between revisions)
| Line 50: | Line 50: | ||
.jumbotron .light-red-text { | .jumbotron .light-red-text { | ||
| - | + | color: #FF7E25; | |
} | } | ||
.jumbotron .red-text { | .jumbotron .red-text { | ||
| - | + | color: #DE4230; | |
} | } | ||
| - | .jumbotron.dark-red-text { | + | .jumbotron .dark-red-text { |
| - | + | color: #AE2726; | |
} | } | ||
| Line 81: | Line 81: | ||
<div class="container"> | <div class="container"> | ||
<a href="https://2014.igem.org" style="position:absolute;z-index:1;"><img src="/wiki/images/5/5d/IGEM_logo_white.png" style="height:70px;" alt="iGEM Logo" /></a> | <a href="https://2014.igem.org" style="position:absolute;z-index:1;"><img src="/wiki/images/5/5d/IGEM_logo_white.png" style="height:70px;" alt="iGEM Logo" /></a> | ||
| - | + | <img id="ring" style="visibility:hidden;" src="/wiki/images/0/0d/Heidelberg_Firering_red.png" alt="Ring of fire Image"/> | |
| - | + | <div class="row"> | |
| - | + | <div class="jumbotron slide epic-background" style="position:relative;"> | |
| - | + | <div id="img-placeholder" class="col-lg-6 col-md-6 col-sm-12"> | |
| - | + | <img id="placeholder" class="img-responsive" src="/wiki/images/0/0d/Heidelberg_Firering_red.png" alt="Ring of fire Image"/> | |
| - | + | </div> | |
| - | + | <div class="col-lg-6 col-md-6 col-sm-12 impressiv"> | |
| - | + | <div class="hidden-sm hidden-xs" style="padding-top:100px;"></div> | |
| - | + | <h3 class="normal-medium-text">iGEM TEAM <span>HEIDELBERG</span> 2014</h3> | |
| - | + | <h1 class="very-large-text">THE RING<br />OF <span>FIRE</span></h1> | |
| - | + | ||
<p> Click <a href="/Team:Heidelberg/Project#Abstract">here</a> to view our abstract. | <p> Click <a href="/Team:Heidelberg/Project#Abstract">here</a> to view our abstract. | ||
<br/> | <br/> | ||
Scroll down to EXPLORE our project. | Scroll down to EXPLORE our project. | ||
| - | + | </div> | |
| - | + | <div class="clearfix"></div> | |
| - | + | <div class="arrow arrow-right"></div > | |
| - | + | <div id="scrollNag" class="arrow arrow-right"></div> | |
| - | + | </div> | |
| - | + | </div> | |
| - | + | <div class="row"> | |
| - | + | <div id="second" class="jumbotron slide grey second"> | |
| - | + | <div id="placeholder-2" class="col-lg-3 col-md-3 hidden-sm hidden-xs"> | |
| - | + | </div> | |
| - | + | <div class="col-lg-9 col-md-9 col-sm-12" style="text-align: right;"> | |
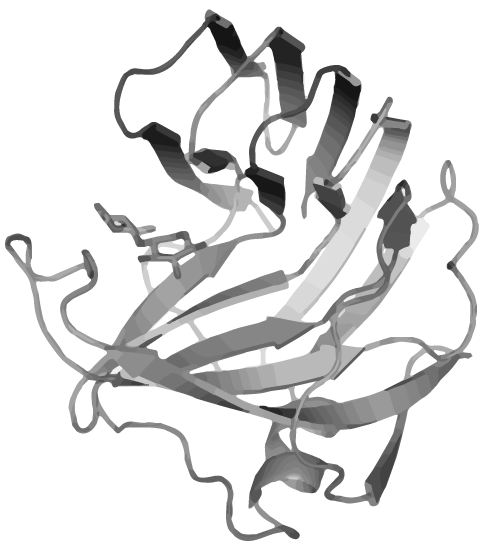
| - | + | <p><span class="larger red-text">Nature</span> has made many curious inventions. One of these are</p> | |
| - | + | <h1 class="very-large-text red-text bold" style="text-align:right;">CIRCULAR PROTEINS</h1> | |
| - | + | <p>which are unconventional peptides that neither have a beginning, nor an ending</p><br> | |
| - | + | <p class="normal-medium-text bold">These proteins are extremely <span class="red-text">resistant</span> against | |
| - | <span class="red-text">high temperatures</span>, | + | <span class="red-text">high temperatures</span>, pH and proteases.</p> |
| - | + | <p class="normal-medium-text align-right">We established protein circularization as a new<br>powerful tool for Synthetic Biology and set the foundations<br>to render any protein heat stable.</p> | |
| - | + | </div> | |
| - | + | <div class="arrow arrow-left"></div> | |
| - | + | <div class="clearfix"></div> | |
| - | + | </div> | |
| - | + | </div> | |
| - | + | <div class="row"> | |
| - | + | <div class="jumbotron slide red red-ring"> | |
| - | + | <h1 class="large-text bold">Wondering how we circularize?</h1> | |
| - | + | <h2 class="normal-medium-text align-right bold">Let us introduce you to the next generation of bioengineering...</h2> | |
| - | + | <h2 class="align-right" style="font-size: 40px;">come DISCOVER the MECHANISM of SPLIT INTEINS</h2><br><br> | |
| - | + | ||
| - | + | <div class="col-lg-6"> | |
| - | + | <p>Inteins excise themselves out of proteins and in doing so the remaining flanking parts are irreversibly joined</p> | |
| - | + | <p>– an effective mechanism to circularize proteins.</p> | |
| - | + | </div> | |
| - | + | <div class="col-lg-6"> | |
| - | + | Placeholder | |
| - | + | </div> | |
| - | + | <div class="arrow arrow-right"></div> | |
| - | + | <div class="clearfix"></div> | |
| - | + | </div> | |
| - | + | </div> | |
| - | + | <div class="row"> | |
| - | + | <div class="jumbotron slide darker-background" style="color:white;"> | |
| - | + | <div> | |
| - | + | <div class="col-xs-12"> | |
| - | + | ||
| - | + | <p class="normal-medium-text align-right">But messing with protein structure can be disastrous,<br><span class="medium-text">you can only win with good MODELING.</span></p><br><br> | |
| - | + | <p class="normal-medium-text"><span>Find out the <span class="bold">EXCITING THEORY</span><br>behind <span class="bold">RIGID LINKERS</span> and their <span class="bold">ANGLES</span></span></p> | |
| - | + | <br> | |
| - | + | <p><span class="larger">We developed CRAUT,<br> | |
| - | + | a comprehensive software which identifies<br> | |
| - | + | the <span class="red-text">optimal path</span> to <span class="red-text">connect</span> a protein’s <span class="red-text">termini</span><br> | |
| - | + | preserving structure and function.</span></p> | |
| - | + | </div> | |
| - | + | <div class="col-lg-12"> | |
| - | + | <p><span class="larger">Look at our extensive wet-lab SCREENING to improve and calibrate our software using <a class="red-text" href="/Team:Heidelberg/Project/Linker_Screening">lambda lysozyme</a>!</span></p> | |
| - | + | </div> | |
| - | + | <div class="clearfix"></div> | |
| - | + | </div> | |
| - | + | <div class="arrow arrow-left"></div> | |
| - | + | </div> | |
| - | + | </div> | |
| - | + | <div class="row"> | |
| - | + | <div class="jumbotron slide grey"> | |
| - | + | <div> | |
| - | + | <div> | |
| - | + | <div class="col-md-4 col-md-offset-0 col-xs-6 col-xs-offset-3" style="z-index:5;"> | |
| - | + | <img class="img-responsive" src="/wiki/images/e/e5/Heidelberg_Frontpage_igemathome.png"> | |
| - | + | </div> | |
| - | + | <div class="col-md-8 col-xs-12"> | |
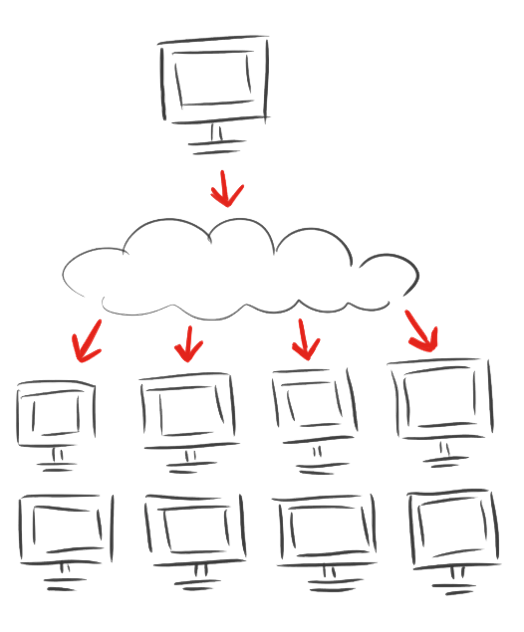
| - | + | <p class="align-right"><span class="larger">As calculations on protein structures are costly<br>we involved the rest of the world with</span></p> | |
| - | + | <h1 class="align-right bold very-large-text dark-red-text" style=" | |
| - | + | color: #AE2726; | |
| - | + | "><span style="font-size:1.5em;">iGEM@home</span></h1> | |
| - | + | </div> | |
| - | + | <div class="clearfix"></div> | |
| - | + | </div> | |
| - | + | <div class="col-md-8 col-xs-12"> | |
| - | + | <h3 class="medium-text bold">Empowering science!</h3><br> | |
| - | + | <p class="normal-medium-text">NEW PLATFORM for distributed computing.</p> | |
| - | + | <p><span class="larger">Volunteers provide the idle calacity of their home computers.</span></p> | |
| - | + | <p class=""><span class="larger">And, with an established user base with more than <span class="larger dark-red-text">1000</span> computers, we effectively bring synthetic biology to society in a new way</span></p> | |
| - | + | </div> | |
| - | + | <div class="col-md-4 col-md-offset-0 col-sm-offset-3 col-sm-6 col-xs-offset-2 col-xs-8"> | |
| - | + | <img style="position:relative; top: -35px;" class="img-responsive" src="/wiki/images/5/52/Heidelberg_Frontpage_igemathome_cloud.png"> | |
| - | + | </div> | |
| - | + | <div class="clearfix"></div> | |
| - | + | </div> | |
| - | + | <div class="arrow arrow-left"></div> | |
| - | + | </div> | |
| - | + | </div> | |
| - | + | <div style="position:relative;" class="row"> | |
| - | + | <div class="jumbotron slide grey"> | |
| - | + | <div> | |
| - | + | <div class="col-md-4 col-md-push-8 col-md-offset-0 col-xs-offset-3 col-xs-6" style="z-index:5;"> | |
| - | + | <img class="img-responsive" src="/wiki/images/a/a6/Heidelberg_Project_Dnmt1.png" /> | |
| - | + | </div> | |
| - | + | <div class="col-md-8 col-md-pull-4 col-xs-12"> | |
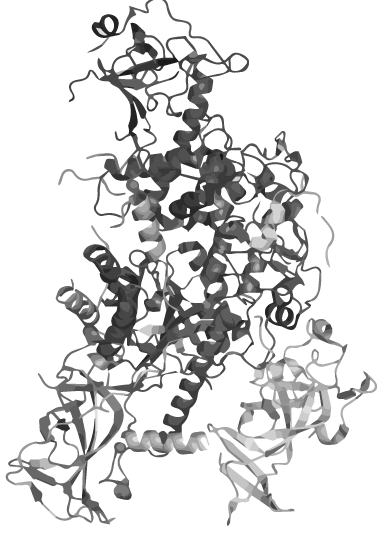
| - | + | <h1 class="dark-grey-text" style="text-align: right;"><span style="font-size: 0.8em;">circular <span class="red-text">heat-stable</span></span><br>DNMT1</h1> | |
| - | + | <p>Wouldn´t it be great to amplify DNA in a normal PCR maintaining the epigenetic information coded in methylation patterns?</p> | |
| - | + | <p>The problem: DNMT 1, an enzyme which is responsible for the establishment and maintenance of the individual methylation pattern of different cell types, is not heat stable. | |
| - | + | For iGEM 2014 we therefore create a PCR 2.0 with heat-stable DNMT 1 by circularization. | |
| - | + | </p> | |
| - | + | </div> | |
| - | + | <div class="clearfix"></div> | |
| - | + | </div> | |
| - | + | <div> | |
| - | + | <div class="title-wrapper-dnmt1"> | |
| - | + | <span class="title-dnmt1">PCR 2.0</span> | |
| - | + | <span class="special-span-dnmt1"></span> | |
| - | + | </div> | |
| - | + | </div> | |
| - | + | <div class="arrow arrow-right"></div> | |
| - | + | </div> | |
| - | + | </div> | |
| - | + | <div class="row"> | |
| - | + | <div class="jumbotron slide epic-background" style="color:white;"> | |
| - | + | <div class="col-lg-4 col-md-4 col-xs-12" style="height: 410px; position: relative"> | |
| - | + | <div id="circ-box" class="descr-box"> | |
| - | + | <a href="/Team:Heidelberg/Toolbox/Circularization"> | |
| - | + | <img src="/wiki/images/5/58/Heidelberg_Toolbox_Circularization.png" id="circ-icon" class="toolbox-icon"> CIRCULARIZATION | |
| - | + | </a> | |
| - | + | </div> | |
| - | + | <div id="oligo-box" class="descr-box"> | |
| - | + | <a href="/Team:Heidelberg/Toolbox/Oligomerization"> | |
| - | + | <img src="/wiki/images/4/40/Oligomerization.png" id="oligo-icon" class="toolbox-icon"> OLIGOMERIZATION | |
| - | + | </a> | |
| - | + | </div> | |
| - | + | <div id="fusion-box" class="descr-box"> | |
| - | + | <a href="/Team:Heidelberg/Toolbox/Fusion"> | |
| - | + | <img src="/wiki/images/8/87/Heidelberg_Toolbox_Fusion.png" id="fusion-icon" class="toolbox-icon"> FUSION | |
| - | + | </a> | |
| - | + | </div> | |
| - | + | <div id="onoff-box" class="descr-box"> | |
| - | + | <a href="/Team:Heidelberg/Toolbox/OnOff"> | |
| - | + | <img src="/wiki/images/c/c2/Heidelberg_Toolbox_On-Off.png" id="onoff-icon" class="toolbox-icon"> ON/OFF | |
| - | + | </a> | |
| - | + | </div> | |
| - | + | <div id="purification-box" class="descr-box"> | |
| - | + | <a href="/Team:Heidelberg/Toolbox/Purification"> | |
| - | + | <img src="/wiki/images/0/04/Heidelberg_Toolbox_Purification.png" id="purification-icon" class="toolbox-icon"> PURIFICATION | |
| - | + | </a> | |
| - | + | </div> | |
| - | + | </div> | |
| - | + | <div class="col-lg-8 col-md-8 col-xs-12"> | |
| - | + | <h1 style="text-align: right; font-size: 3em;" >... and show you the WORLD of <br /> | |
| - | + | <span class="red-text">post-translational MODIFCATION</span> | |
| - | + | </h1> | |
| - | + | <p>The iGEM Team Heidelberg has developed an intein toolbox for the iGEM community to easily modify your protein in a standarized method.</p> | |
| - | + | <p>Our toolbox contains several tools which are PLACEHOLDER. Here you can find out more about our Toolbox.</p> | |
| - | + | <p>In addition to that all tools are inducible by light. Using the LOV system we built a solid method for regulation of the intein trans-splicing reaction our toolbox consiting on. Click here to get more informations about Induction.</p> | |
| - | + | </div> | |
| - | + | <div class="clearfix"></div> | |
| - | + | <div class="arrow arrow-left"></div> | |
| - | + | </div> | |
| - | + | </div> | |
| - | + | <div style="position:relative;" class="row"> | |
| - | + | <div class="jumbotron slide red"> | |
| - | + | <div> | |
| - | + | <div class="col-md-4" style="z-index:5;"> | |
| - | + | <img style="margin-top:25px;"class="img-responsive" src="/wiki/images/8/82/Heidelberg_Project_Xylanase.png" /> | |
| - | + | </div> | |
| - | + | <div class="col-md-8"> | |
| - | + | <h1 style="text-align: right; color: black; padding-right: 130px;" ><span style="font-size: 0.8em;">circular <span style="color:white;">heat-stable</span></span><br><span style="font-size:1.3em;">Xylanase</span></h1> | |
| - | + | <p>Xylanase is an important enzyme for the pulp and paper industry.</p> | |
| - | + | <p>Bla bla</p> | |
| - | + | <p>In future Xylanase could be used for the production of biofuel.</p> | |
| - | + | </div> | |
| - | + | <div class="clearfix"></div> | |
| - | + | </div> | |
| - | + | <div class="title-wrapper-dnmt1" style="text-align: right; color:white; font-weight:bold;"> | |
| - | + | <span class="title-dnmt1">INDUSTRY</span> | |
| - | + | <span class="special-span-dnmt1"></span> | |
| - | + | </div> | |
| - | + | <div class="arrow arrow-left"></div> | |
| - | + | </div> | |
| - | + | </div> | |
| - | + | <div class="row"> | |
| - | + | <div class="jumbotron slide dark-grey" style="color:white; position:relative;"> | |
| - | + | <div class="col-lg-5 col-md-5 hidden-sm hidden-xs" style="position:relative; bottom:-48px;"> | |
| - | + | <img class="img-responsive" src="/wiki/images/5/5e/Heidelberg_Frontpage_Team.png" /> | |
| - | + | </div> | |
| - | + | <div class="col-lg-7 col-md-7 col-sm-12 col-xs-12"> | |
| - | + | <h1>Who are we?</h1> | |
| - | + | <p>We are the iGEM Team Heidelberg 2014 consisting of 12 highly motivated bachelor and master students studying at Heidelberg University.</p> | |
| - | + | <p>For our project we got great feedback and support from our supervisors.</p> | |
| - | + | <p>Take a look at our <a href="/Team:Heidelberg/Team">Teampage</a>!</p> | |
| - | + | <h1>Thank you!</h1> | |
| - | + | <p>We want thank all people who helped us and supported our work in the lab.</p> | |
| - | + | </div> | |
| - | + | ||
| + | <div class="clearfix"></div> | ||
| + | </div> | ||
| + | </div> | ||
</div> | </div> | ||
</html> | </html> | ||
{{:Team:Heidelberg/Templates/IncludeJS|:Team:Heidelberg/js/frontpage}} | {{:Team:Heidelberg/Templates/IncludeJS|:Team:Heidelberg/js/frontpage}} | ||
Revision as of 15:30, 15 October 2014
 "
"



 CIRCULARIZATION
CIRCULARIZATION
 OLIGOMERIZATION
OLIGOMERIZATION
 FUSION
FUSION
 ON/OFF
ON/OFF
 PURIFICATION
PURIFICATION