Team:Heidelberg
From 2014.igem.org
(Difference between revisions)
| Line 21: | Line 21: | ||
} | } | ||
| - | .jumbotron . | + | .jumbotron .medium-large-text { |
font-size: 50px; | font-size: 50px; | ||
} | } | ||
| - | .jumbotron . | + | .jumbotron .medium-text { |
font-size: 42px; | font-size: 42px; | ||
} | } | ||
| - | .jumbotron .normal- | + | .jumbotron .normal-medium-text { |
font-size: 30px; | font-size: 30px; | ||
} | } | ||
| Line 89: | Line 89: | ||
<div class="col-lg-6 col-md-6 col-sm-12 impressiv"> | <div class="col-lg-6 col-md-6 col-sm-12 impressiv"> | ||
<div class="hidden-sm hidden-xs" style="padding-top:100px;"></div> | <div class="hidden-sm hidden-xs" style="padding-top:100px;"></div> | ||
| - | <h3 class="normal- | + | <h3 class="normal-medium-text">iGEM TEAM <span>HEIDELBERG</span> 2014</h3> |
<h1 class="very-large-text">THE RING<br />OF <span>FIRE</span></h1> | <h1 class="very-large-text">THE RING<br />OF <span>FIRE</span></h1> | ||
| Line 109: | Line 109: | ||
<h1 class="very-large-text red-text bold" style="text-align:right;">CIRCULAR PROTEINS</h1> | <h1 class="very-large-text red-text bold" style="text-align:right;">CIRCULAR PROTEINS</h1> | ||
<p>which are unconventional peptides that neither have a beginning, nor an ending</p> | <p>which are unconventional peptides that neither have a beginning, nor an ending</p> | ||
| - | <p class="normal- | + | <p class="normal-medium-text bold">These proteins are extremely <span class="red-text">resistant</span> against |
<span class="red-text">high temperatures</span>, <span class="red-text">pH</span> and <span class="red-text">proteases</span>.</p> | <span class="red-text">high temperatures</span>, <span class="red-text">pH</span> and <span class="red-text">proteases</span>.</p> | ||
| - | <p class="normal- | + | <p class="normal-medium-text align-right">We established protein circularization as a <span class="red-text">new<br/>powerful tool</span> for Synthetic Biology and set the foundations<br />to render any protein heat stable.</p> |
</div> | </div> | ||
<div class="arrow arrow-left"></div> | <div class="arrow arrow-left"></div> | ||
| Line 122: | Line 122: | ||
<div class="jumbotron slide red red-ring"> | <div class="jumbotron slide red red-ring"> | ||
<h1 class="large-text">Wondering how we circularize?</h1> | <h1 class="large-text">Wondering how we circularize?</h1> | ||
| - | <h2 class=" | + | <h2 class="medium-text align-right">Let us introduce you to the next generation of bioengineering...</h2> |
| - | <h2 class=" | + | <h2 class="medium-text align-right">COME DISCOVER THE MECHANISM OF SPLIT INTEINS</h2> |
<div class="col-lg-6"> | <div class="col-lg-6"> | ||
<p>Inteins excise themselves out of proteins and in doing so the remaining flanking parts are irreversibly joined</p> | <p>Inteins excise themselves out of proteins and in doing so the remaining flanking parts are irreversibly joined</p> | ||
| Line 140: | Line 140: | ||
<div class="col-xs-12"> | <div class="col-xs-12"> | ||
<p class="align-right "></p> | <p class="align-right "></p> | ||
| - | <p class="normal- | + | <p class="normal-medium-text">But messing with protein structure can be disastrous,<br/><span class="medium-text">you can only win with good MODELING.</span></p> |
<p>Find out the EXCITING THEORY<br>behind RIGID LINKERS and their ANGLES</p> | <p>Find out the EXCITING THEORY<br>behind RIGID LINKERS and their ANGLES</p> | ||
<br/> | <br/> | ||
| - | <p class="normal- | + | <p class="normal-medium-text">We developed CRAUT,<br/> |
a comprehensive software which identifies<br /> | a comprehensive software which identifies<br /> | ||
the <span class="red-text">optimal path</span> to <span class="red-text">connect</span> a protein’s <span class="red-text">termini</span><br/> | the <span class="red-text">optimal path</span> to <span class="red-text">connect</span> a protein’s <span class="red-text">termini</span><br/> | ||
| Line 250: | Line 250: | ||
</div> | </div> | ||
<div class="col-md-9 col-xs-12"> | <div class="col-md-9 col-xs-12"> | ||
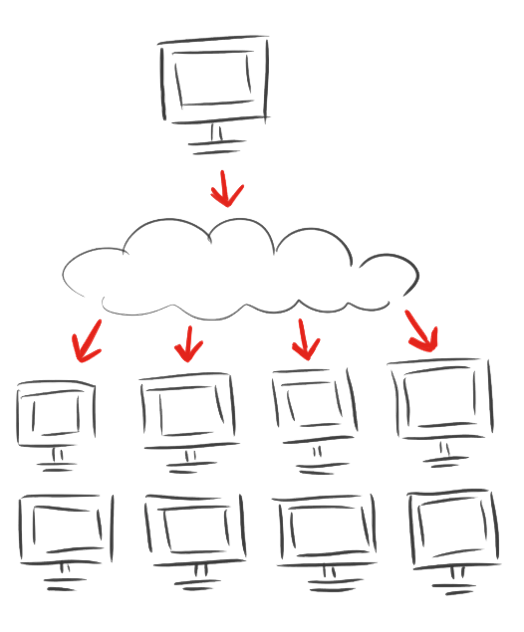
| - | < | + | <p class="larger align-right">As calculations on protein structures are costly<br />we involved the rest of the world with</p> |
| + | <h1 class="very-large-text red-text">iGEM@home</h1> | ||
</div> | </div> | ||
<div class="clearfix"></div> | <div class="clearfix"></div> | ||
</div> | </div> | ||
<div class="col-md-8 col-xs-12"> | <div class="col-md-8 col-xs-12"> | ||
| + | <h3 class="normal-medium-text">Empowering science!</h3> | ||
<p><br/>After calculating for eleven days and the breakdown of both computational and mental power we decided to spread the modeling of the linkers.</p> | <p><br/>After calculating for eleven days and the breakdown of both computational and mental power we decided to spread the modeling of the linkers.</p> | ||
<p>The iGEM Team Heidelberg developed <a href="/Team:Heidelberg/Software/igemathome">iGEM@home</a>, a software to divide extensive computing task into many packages and to distribute them to many computers. Now over 1,000 volunteers are calculating for us when their computers are idle.</p> | <p>The iGEM Team Heidelberg developed <a href="/Team:Heidelberg/Software/igemathome">iGEM@home</a>, a software to divide extensive computing task into many packages and to distribute them to many computers. Now over 1,000 volunteers are calculating for us when their computers are idle.</p> | ||
Revision as of 14:18, 15 October 2014
 "
"


 CIRCULARIZATION
CIRCULARIZATION
 OLIGOMERIZATION
OLIGOMERIZATION
 FUSION
FUSION
 ON/OFF
ON/OFF
 PURIFICATION
PURIFICATION