Team:Evry/Notebook
From 2014.igem.org
| Line 216: | Line 216: | ||
| + | |||
| + | <html> | ||
| + | <head> | ||
| + | |||
| + | <script src="http://ajax.googleapis.com/ajax/libs/jquery/1.11.1/jquery.min.js"></script> | ||
| + | <script> | ||
| + | var main = function() { | ||
| + | $('.icon-menu').click(function() { | ||
| + | $('.menu').animate({ | ||
| + | left: "0px" | ||
| + | }, 200); | ||
| + | |||
| + | $('body').animate({ | ||
| + | left: "285px" | ||
| + | }, 200); | ||
| + | }); | ||
| + | |||
| + | |||
| + | $('.icon-close').click(function() { | ||
| + | $('.menu').animate({ | ||
| + | left: "-285px" | ||
| + | }, 200); | ||
| + | |||
| + | $('body').animate({ | ||
| + | left: "0px" | ||
| + | }, 200); | ||
| + | }); | ||
| + | }; | ||
| + | $(document).ready(main) | ||
| + | |||
| + | </script> | ||
| + | |||
| + | |||
| + | <style> | ||
| + | /* Initial body */ | ||
| + | body { | ||
| + | left: 0; | ||
| + | margin: 0; | ||
| + | overflow: hidden; | ||
| + | position: relative; | ||
| + | } | ||
| + | |||
| + | /* Initial menu */ | ||
| + | .menu { | ||
| + | background: #04084E; | ||
| + | left: -285px; /* start off behind the scenes */ | ||
| + | height: 25%; | ||
| + | position: fixed; | ||
| + | width: 285px; | ||
| + | } | ||
| + | |||
| + | /* Basic styling */ | ||
| + | |||
| + | .jumbotron { | ||
| + | |||
| + | |||
| + | /* background-image: url('/home/wdigan/Desktop/igem/wiki/imagewiki/BernardEvry.JPG'); */ | ||
| + | background: rgb(240,249,255); /* Old browsers */ | ||
| + | background: -moz-linear-gradient(left, rgba(240,249,255,1) 0%, rgba(203,235,255,1) 47%, rgba(161,219,255,1) 100%); /* FF3.6+ */ | ||
| + | background: -webkit-gradient(linear, left top, right top, color-stop(0%,rgba(240,249,255,1)), color-stop(47%,rgba(203,235,255,1)), color-stop(100%,rgba(161,219,255,1))); /* Chrome,Safari4+ */ | ||
| + | background: -webkit-linear-gradient(left, rgba(240,249,255,1) 0%,rgba(203,235,255,1) 47%,rgba(161,219,255,1) 100%); /* Chrome10+,Safari5.1+ */ | ||
| + | background: -o-linear-gradient(left, rgba(240,249,255,1) 0%,rgba(203,235,255,1) 47%,rgba(161,219,255,1) 100%); /* Opera 11.10+ */ | ||
| + | background: -ms-linear-gradient(left, rgba(240,249,255,1) 0%,rgba(203,235,255,1) 47%,rgba(161,219,255,1) 100%); /* IE10+ */ | ||
| + | background: linear-gradient(to right, rgba(240,249,255,1) 0%,rgba(203,235,255,1) 47%,rgba(161,219,255,1) 100%); /* W3C */ | ||
| + | filter: progid:DXImageTransform.Microsoft.gradient( startColorstr='#f0f9ff', endColorstr='#a1dbff',GradientType=1 ); /* IE6-9 */ | ||
| + | height: 100%; | ||
| + | -webkit-background-size: cover; | ||
| + | -moz-background-size: cover; | ||
| + | -o-background-size: cover; | ||
| + | background-size: cover; | ||
| + | } | ||
| + | |||
| + | .menu ul { | ||
| + | border-top: 4px solid #636366; | ||
| + | list-style: none; | ||
| + | margin: 0; | ||
| + | padding: 0; | ||
| + | } | ||
| + | |||
| + | .menu li { | ||
| + | border-bottom: 1px solid #636366; | ||
| + | font-family: 'Open Sans', sans-serif; | ||
| + | line-height: 20px; | ||
| + | padding-bottom: 3px; | ||
| + | padding-left: 20px; | ||
| + | padding-top: 3px; | ||
| + | } | ||
| + | |||
| + | /*icon tab*/ | ||
| + | .menu a { | ||
| + | color: white; | ||
| + | font-size: 15px; | ||
| + | text-decoration: none; | ||
| + | text-transform: uppercase; | ||
| + | } | ||
| + | |||
| + | .icon-close { | ||
| + | cursor: pointer; | ||
| + | padding-left:5px; | ||
| + | padding-top: 5px; | ||
| + | } | ||
| + | |||
| + | /*contents tab*/ | ||
| + | .icon-menu { | ||
| + | color: black; | ||
| + | cursor: pointer; | ||
| + | font-family: 'Open Sans', sans-serif; | ||
| + | font-size: 25px; | ||
| + | padding-bottom: 25px; | ||
| + | padding-left: 25px; | ||
| + | padding-top: 25px; | ||
| + | text-decoration: none; | ||
| + | text-transform: uppercase; | ||
| + | } | ||
| + | |||
| + | .icon-menu i { | ||
| + | margin-right: 5px; | ||
| + | } | ||
| + | |||
| + | </style> | ||
| + | </head> | ||
| + | <body> | ||
| + | |||
| + | <div class="menu"> | ||
| + | |||
| + | <!-- Menu icon --> | ||
| + | <div class="icon-close"> | ||
| + | <img src="http://s3.amazonaws.com/codecademy-content/courses/ltp2/img/uber/close.png"> | ||
| + | </div> | ||
| + | |||
| + | <!-- Menu --> | ||
| + | <ul> | ||
| + | <li><a href="#">Cell characterization</a></li> | ||
| + | <li><a href="#">Transformation</a></li> | ||
| + | <li><a href="#">RNA sequencing</a></li> | ||
| + | <li><a href="#">Sensing</a></li> | ||
| + | <li><a href="#">Interlabstudy</a></li> | ||
| + | <li><a href="#">Protocols</a></li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <!-- Main body --> | ||
| + | <div class="jumbotron"> | ||
| + | |||
| + | <div class="icon-menu"> | ||
| + | <i class="fa fa-bars"></i> | ||
| + | Contents | ||
| + | </div> | ||
| + | |||
| + | |||
| + | </body> | ||
| + | </html> | ||
Revision as of 08:45, 19 August 2014

Notebook
Week 7
Interlab Study
08.11.2014
08.12.2014
The 4 needed parts are: BBa_J23101, BBa_J23115, BBa_E0240 and BBa_I20260. Corresponding wells were located on 2014 Distribution kit plates and resuspended with 10 µL steril water. That permits to obtain a DNA concentration around 0.2 ng/µl (according to the registry. Solutions were transferred into 1 ml eppendorf tubes and stored at -20°C.
To amplify fragments, a PCR was performed on the 4 constructions, with the mix described on table 1.
Distribution of 49 µl of mix per PCR tube. Application of program IGEM Q5 PCR.
Preparation of a 1% agarose gel: 0.56 g of Top Vision agarose (Thermo Scientific) + 50 ml of TAE 1X.Microwave 30s by 30s until agarose total dissolution. Gel was cooling down until to be lukewarm, one BET drop was added. Gel was loaded with 10µl per sample previously added with 2 µl of loading dye 6X, and 5 µl for ladders. Gel running 45 minutes at 100 mV in TAE 1X buffer.
We expected to obtain one band per PCR sample corresponding to the interesting amplified fragment. For Lane 2, 3 and 6 it was ok. We had the expected profile. By contrast 2 bands were visible, one at the expected size (around 1200 bp) and another around 600 bp. We decided to perform a purification on gel of each bands.
08.13.2014
PCR products were cleaned with the GeneJET purification kit (Thermo Scientific)followed by DNA quantification with the NanoDrop 2000 (Thermo Scientific).
Preparation of a 1% agarose gel: 0.56 g of Top Vision agarose (Thermo Scientific) + 50 ml of TAE 1X. Microwave 30s by 30s until agarose total dissolution Gel was cooling down until to be lukewarm, one BET drop was added. Gel was loaded with 10µl per sample previously added with 2 µl of loading dye 6X, and 5 µl for ladders. Gel running 45 minutes at 100 mV in TAE 1X buffer.
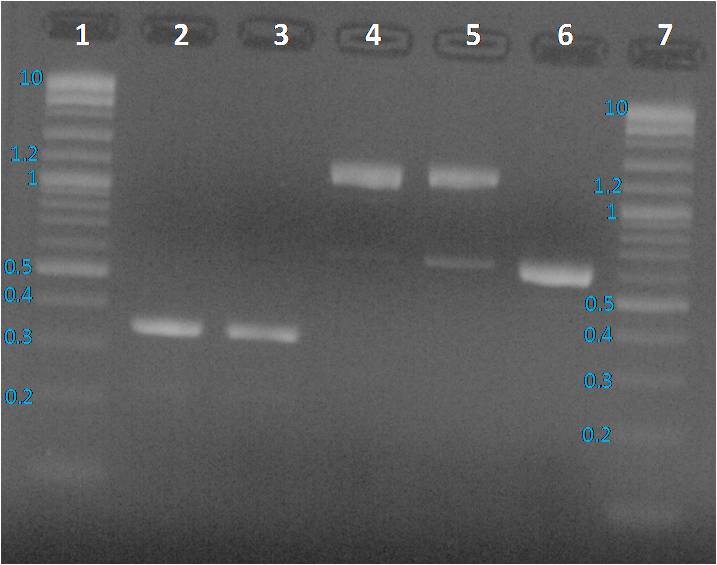
Profiles were same as before PCR clean up. To sequence the amplified parts, we decided to a purification of fragments from the gel as presented below.
Preparation of a 1% agarose gel: 0.56 g of Top Vision agarose (Thermo Scientific) + 50 ml of TAE 1X. Microwave 30s by 30s until agarose total dissolution. Gel was cooling down until to be lukewarm, one BET drop was added. Gel was loaded with 30 µl of BBa_E0240 purified PCR product and BBa_I20260 purified PCR product added with 6 µl of loading dye, at 10µl of the mix per well. Gel running 45 minutes at 100 mV in TAE 1X buffer.
Four pieces of gel were sampled in four tubes to perform a DNA purification from gel.
08.14.2014
To amplify BBa_I20260 and BBa_0240, a PCR was perform with the mix described Table 1. The program IGEM Q5 PCR was applied, see Table 2. PCR products were cleaned with the GeneJET purification kit (Thermo Scientific)followed by DNA quantification with the NanoDrop 2000 (Thermo Scientific).
Preparation of a 1% agarose gel: 0.52 g of Top Vision agarose (Thermo Scientific) + 50 ml of TAE 1X. Microwave 30s by 30s until agarose total dissolution Gel was cooling down until to be lukewarm, one BET drop was added. Gel was loaded with 20 µl of BBa_E0240 PCR product and BBa_I20260 purified PCR product added with 4 µl of loading dye, at 10µl of the mix per well.
Gel running 45 minutes at 100 mV in TAE 1X buffer.
3 pieces of gel were sampled in four tubes to perform a DNA purification from gel. The DNA purification was made with the GeneJET purification kit (Thermo Scientific.
A verification electrophoresis was performed on a 1% agarose gel.
The major part of the DNA was lost during the purification because the amount of was to weak to be purified from gel. So we decide do transform ''E. coli'' to isolate a colony containing the desired plasmid for BBa_E0240 and BBa_I20260.
08.15.2014
BBa_E0240 and BBa_I20260 are respectively into a PSB1C3 and a PSB1K3 vector. In order to select colonies, LB complemented with kanamycin or chloramphenicol are necessary. 200 ml of LB agar medium was prepared with 7 g of LB Agar powder(Sigma) into 200 ml of miliQ water. After complete dissolution, the solution was sterilized in the Tuttmauer 2540ML apparatus, STE 20 minutes at 121°C and EXT+DRY 15 minutes. 50 µl of kanamycin stock solution (25 mg/L) was added to 50 ml of LB agar to pour 2 plates. 150 µl of chloramphenicol stock solution was added in the 150 LB agar remaining ml to pour 6 plates.
The transformation was performed on DH5 alpha ''E. coli'', as followed:
- Remove E. coli competent tubes from -80°C and keep it on ice
- Add 1 µl of template (here solubilized plasmids from the registry distribution kit) and mix gently
- Incubate 10 minutes on ice
- Perform an heat shock 30 seconds at 42°C
- Incubate 2 minutes on ice
- Add 3 ml of LB medium and incubate 60 minutes at 37°C with an agitation at 200 rpm
- Plate 200 µl of BBa_E0240 on a chloramphenicol LB agar plate and BBa_I20260 on a Knamycin LB agar plate
- Incubate plate overnight at 37°C
08.16.2014
Nothing had grown during the night on both plates. Taking into consideration different hypothesis, the transformation was repeated with a change on step 6 and 7. At step 6, only 1 ml of LB medium was added. To plate, 200 µl of BBa_E0240 and BBa_I20260 were plated on 3 plates: LB agar, LB agar kanamycin and LB agar chloramphenicol.
08.17.2014
Colonies had grown on plates as followed:- LB agar: more than 1000 colonies for the two biobricks
- LB agar kanamycin: 10 colonies for BBa_E0240 and >40 colonies for BBa_I20260
- LB agar chloramphenicol: >50 colonies for BBa_E0240 and 3 colonies for BBa_I20260
Plate storage at 4°C.
 "
"






