Team:ETH Zurich/project/overview/summarysimple
From 2014.igem.org
(Difference between revisions)
(→We are driven by this fascination and try to reproduce these snail shells patterns, called Sierpinski triangles, on our own grid of cells.) |
(/* We are driven by this fascination and try to reproduce these snail shells patterns, called Sierpinski triangles, on our own grid of bacteria. To achieve this goal, we follow a synthetic biology approach : we implement the rule that leads to emerge) |
||
| Line 12: | Line 12: | ||
<br/> | <br/> | ||
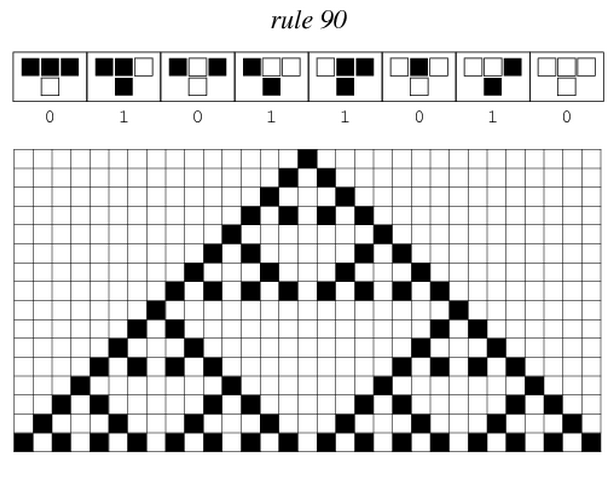
| - | ===== We are driven by | + | ===== We are driven by our fascination for the emergence of complexity and try to make these snail shells patterns, called Sierpinski triangles, emerge on our own grid of bacteria. To achieve this goal, we follow a synthetic biology approach : we implement the rule that leads to emergence of these patterns in the DNA of these bacteria.===== |
===== This way, Mosaicoli not only investigates how a complex pattern can emerge from simple rules, but also develops new tools for controlling communication between cells and implementing reliable biological circuits. ===== | ===== This way, Mosaicoli not only investigates how a complex pattern can emerge from simple rules, but also develops new tools for controlling communication between cells and implementing reliable biological circuits. ===== | ||
Revision as of 12:28, 2 September 2014
You must be wondering where these patterns on snea snail shells come from. What if they would come from a simple rule, followed by all cells on the shell ?
As a matter of fact, many of the complex patterns you can see in nature come from simple rules. It is the case for hurricanes, flocks of birds, neural networks... We call this phenomenon emergence. Emergent phenomena are not predictable from the initial situation and this is why they surprise us. If you are interested in emergence of complexity in general, you can read more about the background of our project.
 "
"