Team:ETH Zurich/project/infopro
From 2014.igem.org
(→Why we chose this track) |
(→Sending) |
||
| (3 intermediate revisions not shown) | |||
| Line 15: | Line 15: | ||
=== The goal: Emergence of patterns via information processing=== | === The goal: Emergence of patterns via information processing=== | ||
| - | We implement a cellular automaton with bacterial cells containing our logic circuitry. Each bacterial colony serves as a core, computing an XOR gate. First, a sensor device detects the input, ''N''-acyl homoserine lactones (AHL). Then, the cell integrates this signal through a logic gate, performed by serine integrases by sensing on the protein level and acting on the DNA level. A necessary post-processing step allows then the production of a new AHL variant due to activated gene expression through the integrase. Meanwhile, | + | We implement a cellular automaton with bacterial cells containing our logic circuitry. Each bacterial colony serves as a core, computing an XOR gate. First, a sensor device detects the input, ''N''-acyl homoserine lactones (AHL). Check the [https://2014.igem.org/Team:ETH_Zurich/project/background/biotools#Biological_Tools quorum sensing and integrases article] for more information. Then, the cell integrates this signal through a logic gate, performed by serine integrases by sensing on the protein level and acting on the DNA level. A necessary post-processing step allows then the production of a new AHL variant due to activated gene expression through the integrase. Meanwhile, green fluorescent protein (GFP) indicates the state of the colony and serves as a long-lasting visual read out. The produced AHL output-signal then propagates in a directed fashion through a [https://2014.igem.org/Team:ETH_Zurich/lab/chip millifluidic] grid to the next bacterial colony. This iterative process faces the challenges of [https://2014.igem.org/Team:ETH_Zurich/expresults/rr#Riboregulators leakiness], crosstalk, protein-level computation and exact [https://2014.igem.org/Team:ETH_Zurich/modeling/diffmodel diffusion] steps. This information pathway is shown in figure 1. |
| - | + | ||
| - | + | ||
| - | + | ||
| + | [[File:ETH_Zurich2014_info_processing1.png|center|800px|thumb|'''Figure 1''' The information pathway in the project Mosai''coli'' on a colony level. Each cell colony is part of a greater memory unit (left-hand side) and gets its input information from the neighboring colonies. The input is given in the form of two different AHLs, here named A and B. This information is then step-wise processed in each of the colonies: sensing - computing - producing - sending. The output molecules serve then as the input for the next colony (middle and left-hand side). This iterative process allows the information processing from the top row of the memory unit to the bottom by chemical wiring via diffusing signals and bio-computations inside of the colonies.]] | ||
=== The steps involved: From sensing to sending=== | === The steps involved: From sensing to sending=== | ||
| Line 48: | Line 46: | ||
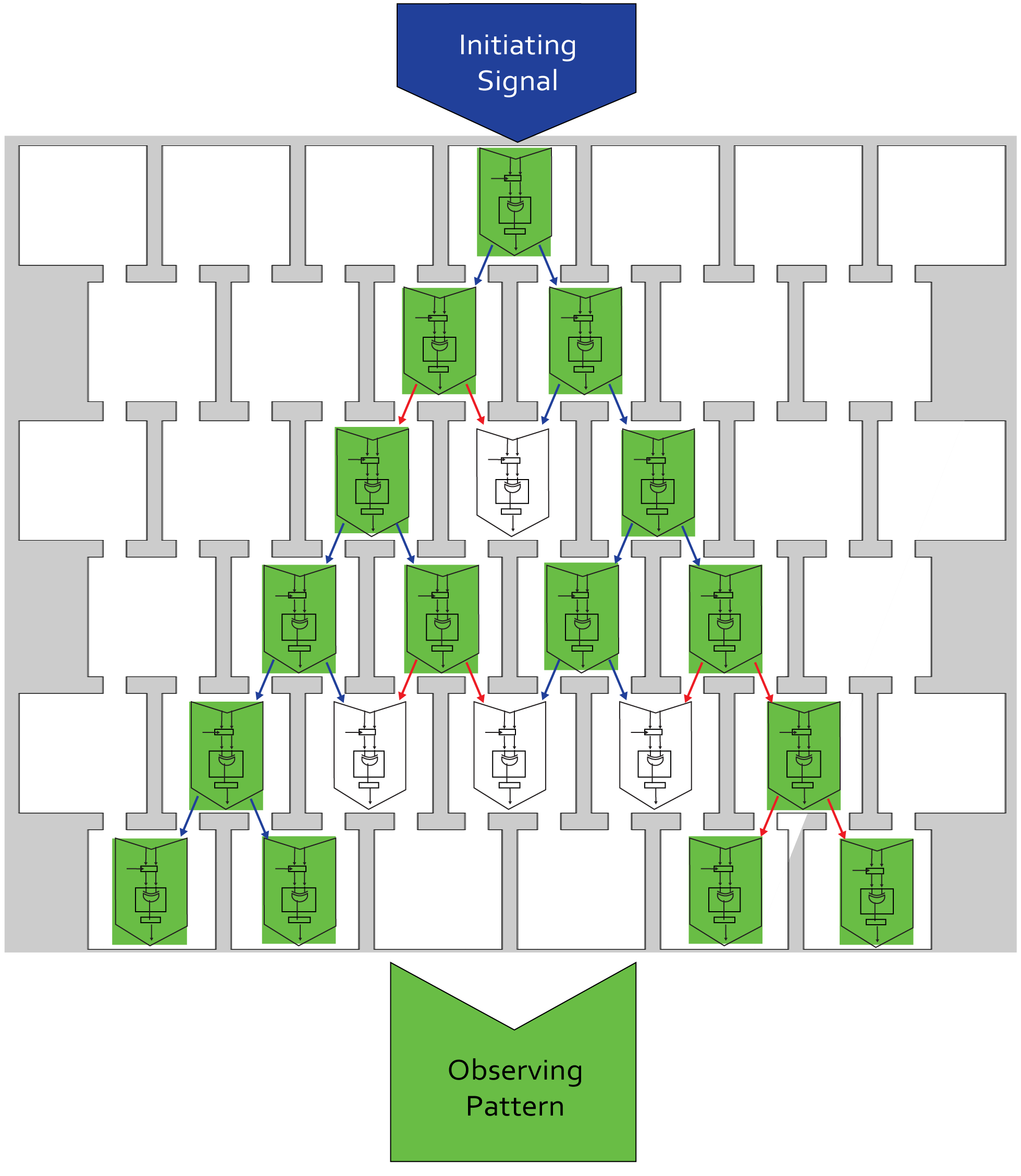
| - | [[File:ETH_Zurich_Information_Processing_chip.png|center|600px|thumb|'''Figure 2 The information pathway in the project Mosai''coli'' on the chip level. | + | [[File:ETH_Zurich_Information_Processing_chip.png|center|600px|thumb|'''Figure 2''' The information pathway in the project Mosai''coli'' on the chip level. After initializing the signal in the top row, it propagates through the wells chemically wired by directed diffusion. In each well, a bacterial colony has to be able to proceed in the information pathway: sensing, integrating (computing), producing and sending. These successive iterations leads to possible error propagation and robustness is one major issue of our system.]] |
<html></article></html> | <html></article></html> | ||
{{:Team:ETH_Zurich/tpl/foot}} | {{:Team:ETH_Zurich/tpl/foot}} | ||
Latest revision as of 03:38, 18 October 2014
Information Processing
Information Processing
About the track
Why we chose this track
With our project, Mosaicoli, we investigate the emergence of complex patterns from simple mathematical rules. Such rules can be reduced to Boolean logic gates, in our case XOR gates. The computations were implemented with integrases, proteins modifying DNA between specific sites. These modifications in turn influence the expression of other proteins which can then indicate the previous change on the genetic level. Additionally chemical wiring of the information to the next cells happens, this again on the protein or small-molecule level. This wiring of information and repeated information processing allows the construction of cellular automata and eventually biocomputers using the analogy of electrical circuits.
The goal: Emergence of patterns via information processing
We implement a cellular automaton with bacterial cells containing our logic circuitry. Each bacterial colony serves as a core, computing an XOR gate. First, a sensor device detects the input, N-acyl homoserine lactones (AHL). Check the quorum sensing and integrases article for more information. Then, the cell integrates this signal through a logic gate, performed by serine integrases by sensing on the protein level and acting on the DNA level. A necessary post-processing step allows then the production of a new AHL variant due to activated gene expression through the integrase. Meanwhile, green fluorescent protein (GFP) indicates the state of the colony and serves as a long-lasting visual read out. The produced AHL output-signal then propagates in a directed fashion through a millifluidic grid to the next bacterial colony. This iterative process faces the challenges of leakiness, crosstalk, protein-level computation and exact diffusion steps. This information pathway is shown in figure 1.
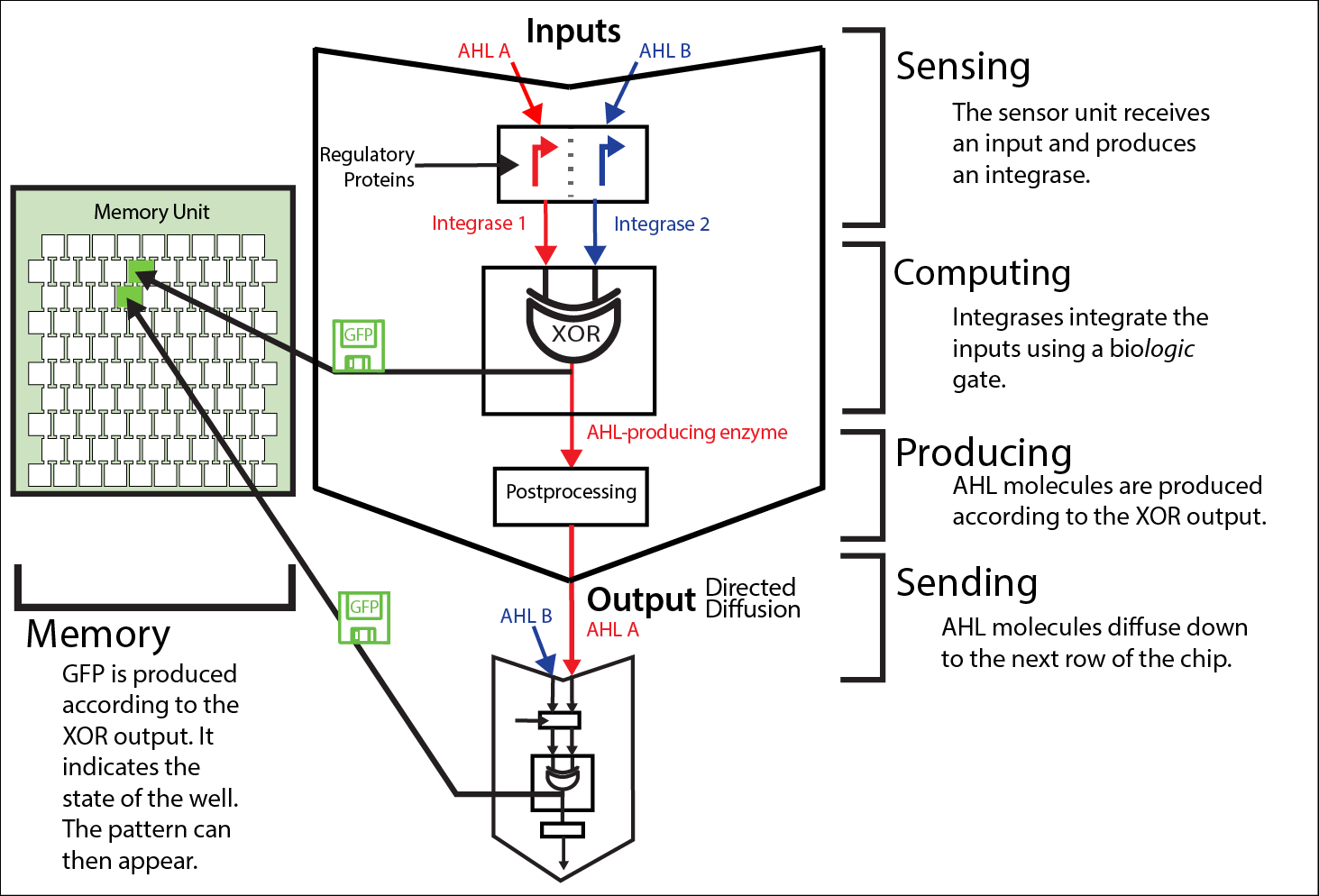
The steps involved: From sensing to sending
Sensing
Diffused AHL molecules are identified via the quorum sensing machinery. The transfer function between the sensed AHL concentration and the promoter activation deviates usually from the ideal one due to leakiness and crosstalk between different types of AHL molecules, regulatory proteins and promoters (we characterized it for Lux promoter (BBa_R0062), Las promoter (BBa_R0079) and Rhl promoter (BBa_R0071)). To limit the risk of error propagation, we implemented a riboregulated system.
Computing
Integrases compute an XOR logic gate via single switching or double switching of a terminator. Their behavior depends on a set of parameters.
Producing
To propagate the computational results, the output has to be translated again into a quorum sensing molecule. Therefore a synthase is expressed to catalyze the production of a new AHL signal.
Memorising
Alongside the AHL synthase, GFP is produced according to the XOR truth table. The fluorescent protein acts both as long term memory for the colonies in ON-state and as visual signal, storing and showing the propagating pattern formation.
Sending
The produced AHL diffuses through channels on the memory unit to the rows below, becoming the input of the following information processing layer. The speed of this 'wiring' by directed diffusion determines the time needed for the pattern to emerge (as long as the channels are relatively long and diffusion is the time-limting step as compared to the actual computation itself). You can find out more information about diffusion and chip design on our modeling page, our experimental results page and our chip page.
The overall process is summarized for the chip, or memory unit, level in figure 2.
 "
"













