Team:ETH Zurich/project/background/biotools
From 2014.igem.org
(→Quorum Sensing) |
(→Integrases) |
||
| (9 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
== Biological Tools == | == Biological Tools == | ||
| - | === Quorum Sensing === | + | === '''Quorum Sensing''' === |
Bacteria, usually found in multi-species neighborhoods with billions of cells, where thought to live and divide isolated from its surroundings. However, bacteria do not try to out-compete the neighbors by mindless exponential growth but can exchange information via cell-to-cell communications in order to coordinate gene-expression and as a result the overall behavior of the population. This communicative endeavor is commonly described as quorum sensing and based on the synthesis, secretion and diffusion of small molecules or 'autoinducers'. | Bacteria, usually found in multi-species neighborhoods with billions of cells, where thought to live and divide isolated from its surroundings. However, bacteria do not try to out-compete the neighbors by mindless exponential growth but can exchange information via cell-to-cell communications in order to coordinate gene-expression and as a result the overall behavior of the population. This communicative endeavor is commonly described as quorum sensing and based on the synthesis, secretion and diffusion of small molecules or 'autoinducers'. | ||
| Line 7: | Line 7: | ||
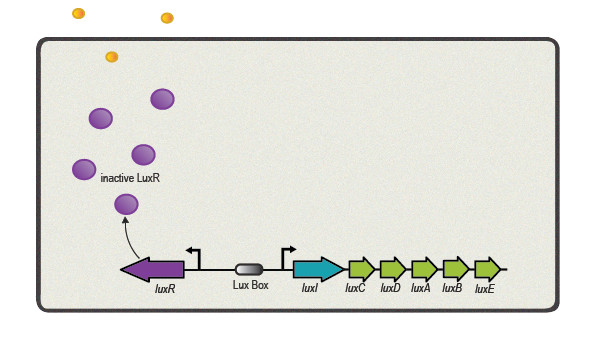
One of the best studied quorum sensing models was found in the Gram-negative bacterium ''V. fischeri'' in the early 1980s<sup>[[Team:ETH_Zurich/project/references#refEberhard|[4]]]</sup>. Depending on the cell density and the corresponding autoinducer concentration, communication between the bacteria induces bioluminescence in the whole ''V. fischeri'' population. The small molecule responsible for this behavior was identified as ''N''-(3-oxohexanoyl)-L-homoserine lactone, which is part of the ''N''-acyl homoserine lactone (AHL) family. This AHL is synthesized by an enzyme called LuxI, diffuses through cell membranes and can then reach the cytosol of neighboring cells. There it binds to and activates LuxR, a protein positively regulating gene transcription. Subsequently to the investigation of ''V. fischeri'', several other AHLs (with specific acyl side chains) and corresponding LuxR/LuxI like systems were found in various bacterial species, coordinating diverse responses including plasmid transfer, virulence, and antibiotic resistance<sup>[[Team:ETH_Zurich/project/references#refWilliams|[5]]]</sup>. | One of the best studied quorum sensing models was found in the Gram-negative bacterium ''V. fischeri'' in the early 1980s<sup>[[Team:ETH_Zurich/project/references#refEberhard|[4]]]</sup>. Depending on the cell density and the corresponding autoinducer concentration, communication between the bacteria induces bioluminescence in the whole ''V. fischeri'' population. The small molecule responsible for this behavior was identified as ''N''-(3-oxohexanoyl)-L-homoserine lactone, which is part of the ''N''-acyl homoserine lactone (AHL) family. This AHL is synthesized by an enzyme called LuxI, diffuses through cell membranes and can then reach the cytosol of neighboring cells. There it binds to and activates LuxR, a protein positively regulating gene transcription. Subsequently to the investigation of ''V. fischeri'', several other AHLs (with specific acyl side chains) and corresponding LuxR/LuxI like systems were found in various bacterial species, coordinating diverse responses including plasmid transfer, virulence, and antibiotic resistance<sup>[[Team:ETH_Zurich/project/references#refWilliams|[5]]]</sup>. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | [[File:ETH Zurich 2014_LuxBoxLowdensity.png|center|500px|thumb|'''Figure 4''' Lux Low Density]] | |
| - | + | ||
| - | + | [[File:EHT Zurich 2014_LuxBoxHighDensity.png|center|500px|thumb|'''Figure 5''' Lux High Density]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | [[File:ETH Zurich 2014_LuxBoxLowdensity.png|center|500px|thumb|Lux Low Density]] | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | [[File:EHT Zurich 2014_LuxBoxHighDensity.png|center|500px|thumb|Lux High Density]] | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Here we focus in particular on the quorum sensing circuitry of ''V. fischeri'' and ''Pseudomonas aeruginosa'', another Gram-negative bacteria. The additional systems are called LasR/LasI and RhlR/RhlI, also employing AHL molecules as autoinducers and are similar to the prototypical LuxR/LuxI system. Even though a key feature of quorum sensing molecules and its receptors is high specificity designed for intraspecies communication, the structural similarity of the different AHL molecules can cause unwanted gene expression. This cross-talk has to be taken into account when re-engineering LuxR/LuxI like systems for synthetic modules and circuits<sup>[[Team:ETH_Zurich/project/references#refJayaraman|[6]]]</sup>. | Here we focus in particular on the quorum sensing circuitry of ''V. fischeri'' and ''Pseudomonas aeruginosa'', another Gram-negative bacteria. The additional systems are called LasR/LasI and RhlR/RhlI, also employing AHL molecules as autoinducers and are similar to the prototypical LuxR/LuxI system. Even though a key feature of quorum sensing molecules and its receptors is high specificity designed for intraspecies communication, the structural similarity of the different AHL molecules can cause unwanted gene expression. This cross-talk has to be taken into account when re-engineering LuxR/LuxI like systems for synthetic modules and circuits<sup>[[Team:ETH_Zurich/project/references#refJayaraman|[6]]]</sup>. | ||
<br> <br> | <br> <br> | ||
| - | === Integrases === | + | === '''Integrases''' === |
Site specific serine recombinases, or integrases, are enzymes recognizing pairs of short and non-identical DNA sequences. Within those sites they irreversibly catalyze the excision or unidirectional inversion of the DNA bases, depending on the orientation of the recognition sequence (often referred to as attB or attP). Many recombinases originate from phages integrating their genes into bacterial host genomes, hence they often do not require specific co-factors and are fully functional when heterologously expressed in bacteria. As a result, they can be employed as molecular tools in biotechnology<sup>[[Team:ETH_Zurich/project/references#refKobi|[7]]]</sup>. | Site specific serine recombinases, or integrases, are enzymes recognizing pairs of short and non-identical DNA sequences. Within those sites they irreversibly catalyze the excision or unidirectional inversion of the DNA bases, depending on the orientation of the recognition sequence (often referred to as attB or attP). Many recombinases originate from phages integrating their genes into bacterial host genomes, hence they often do not require specific co-factors and are fully functional when heterologously expressed in bacteria. As a result, they can be employed as molecular tools in biotechnology<sup>[[Team:ETH_Zurich/project/references#refKobi|[7]]]</sup>. | ||
| Line 78: | Line 24: | ||
| - | [[File:SerineRecombinases.png|center|700px|thumb|Serine Recombinases<sup>[[Team:ETH_Zurich/project/references#refBrown|[ | + | [[File:SerineRecombinases.png|center|700px|thumb|'''Figure 6''' Serine Recombinases<sup>[[Team:ETH_Zurich/project/references#refBrown|[14]]]</sup>.]] |
Latest revision as of 02:31, 18 October 2014
Biological Tools
Quorum Sensing
Bacteria, usually found in multi-species neighborhoods with billions of cells, where thought to live and divide isolated from its surroundings. However, bacteria do not try to out-compete the neighbors by mindless exponential growth but can exchange information via cell-to-cell communications in order to coordinate gene-expression and as a result the overall behavior of the population. This communicative endeavor is commonly described as quorum sensing and based on the synthesis, secretion and diffusion of small molecules or 'autoinducers'.
One of the best studied quorum sensing models was found in the Gram-negative bacterium V. fischeri in the early 1980s[4]. Depending on the cell density and the corresponding autoinducer concentration, communication between the bacteria induces bioluminescence in the whole V. fischeri population. The small molecule responsible for this behavior was identified as N-(3-oxohexanoyl)-L-homoserine lactone, which is part of the N-acyl homoserine lactone (AHL) family. This AHL is synthesized by an enzyme called LuxI, diffuses through cell membranes and can then reach the cytosol of neighboring cells. There it binds to and activates LuxR, a protein positively regulating gene transcription. Subsequently to the investigation of V. fischeri, several other AHLs (with specific acyl side chains) and corresponding LuxR/LuxI like systems were found in various bacterial species, coordinating diverse responses including plasmid transfer, virulence, and antibiotic resistance[5].
Here we focus in particular on the quorum sensing circuitry of V. fischeri and Pseudomonas aeruginosa, another Gram-negative bacteria. The additional systems are called LasR/LasI and RhlR/RhlI, also employing AHL molecules as autoinducers and are similar to the prototypical LuxR/LuxI system. Even though a key feature of quorum sensing molecules and its receptors is high specificity designed for intraspecies communication, the structural similarity of the different AHL molecules can cause unwanted gene expression. This cross-talk has to be taken into account when re-engineering LuxR/LuxI like systems for synthetic modules and circuits[6].
Integrases
Site specific serine recombinases, or integrases, are enzymes recognizing pairs of short and non-identical DNA sequences. Within those sites they irreversibly catalyze the excision or unidirectional inversion of the DNA bases, depending on the orientation of the recognition sequence (often referred to as attB or attP). Many recombinases originate from phages integrating their genes into bacterial host genomes, hence they often do not require specific co-factors and are fully functional when heterologously expressed in bacteria. As a result, they can be employed as molecular tools in biotechnology[7].
Due to the site-specificity, recombinases like Bxb1 and ΦC31 can be employed orthogonally in synthetic systems. In addition, the combination of multiple recombinases -and their corresponding specific recognition sites- with gene regulatory sequences like promoters and terminators, allowed the engineering of Boolean logic gates[8]. For example, when an asymmetric terminator sequence is placed within a pair of recombination sites, the RNA polymerase can only proceed along this sequence if exactly one recombinase is available. Otherwise the terminator is blocking gene transription by default, or due to the repeated inversion, is again fully functional as soon as both recombinases are present[9]. Such recombinase-based logic gates are at the core of our genetic circuits.

 "
"