Team:ETH Zurich/labblog/20140705
From 2014.igem.org
| (28 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | <html>< | + | <html><article class="mix lab plasmids carousel" id='constructionreg' date="20140705"></html> |
| - | = | + | |
| - | + | ||
| - | + | ||
| - | + | == Construction of the regulator plasmids == | |
| - | = | + | ==== Thursday, July 5th ==== |
| - | + | ||
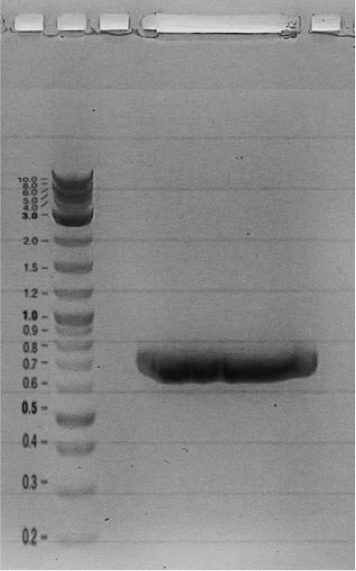
| - | <html></ | + | === Construction of the ''lasR'' regulator plasmid (piG0040) === |
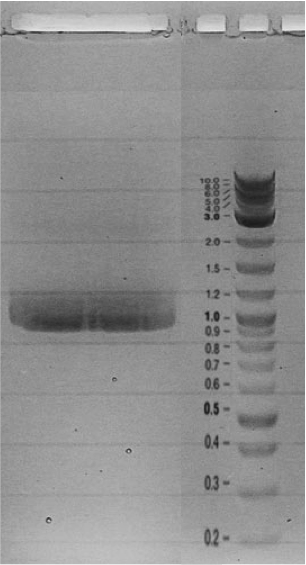
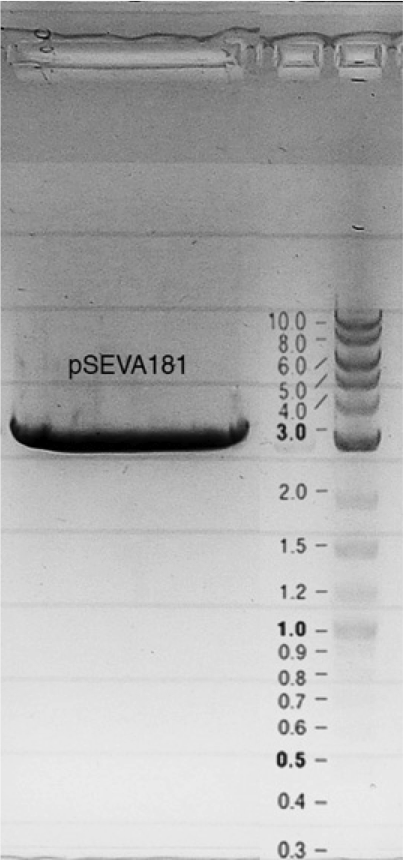
| + | Competent cells were transformed with piG0028 (K553003, ''lasR'') and selected on chloramphenicol-LB-plates. Plasmid DNA was extracted by performing a miniprep. The relevant plasmid sequence was sequenced by Microsynth using the primers oiG0001 and oiG0002. Amplification of ''lasR'' by PCR using oiG0003 and oiG0004 resulted in the fragment fiG0004 (1.0 kb). The vector piG0034 (pSEVA 181) was digested with the restriction enzymes HindIII and PacI to fiG0001 (3.0 kb). The two fragments, fiG0001 and fiG0004, were assembled using Gibbson assembly. Thus we used the plasmid backbone of piG0034 and ''lasR'' of piG0028 to construct the ''lasR'' regulator plasmid piG0040. Competent cells were transformed with piG0040 and selected on ampicillin-LB-plates. Colony PCR using the primers oiG0035 and oiG0036 was conducted to check the size of the inserted fragment (expected bands at 1.0 and 1.2 kb). The sequence was verified by Microsynth using the same primers. | ||
| + | |||
| + | [[File:f4.png|x300px]] | ||
| + | [[File:pSEVA181.png|x300px]] | ||
| + | [[File:20140710_GA_colony_PCR.jpg|x300px]] | ||
| + | |||
| + | |||
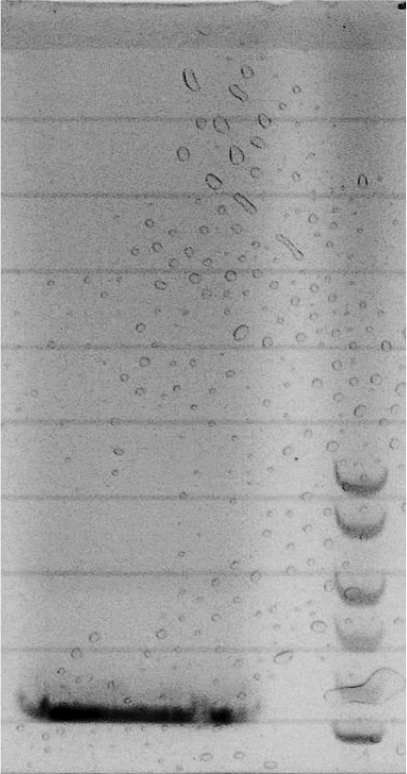
| + | ===Construction of the ''luxR'' regulator plasmid (piG0041)=== | ||
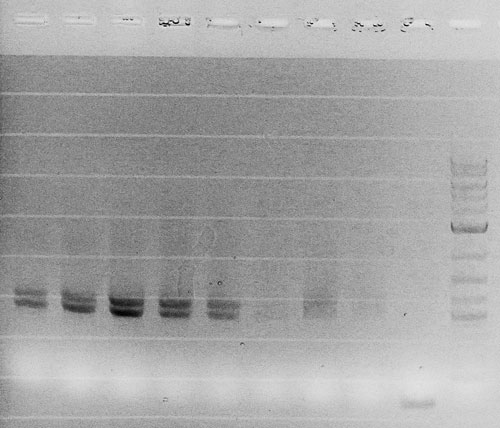
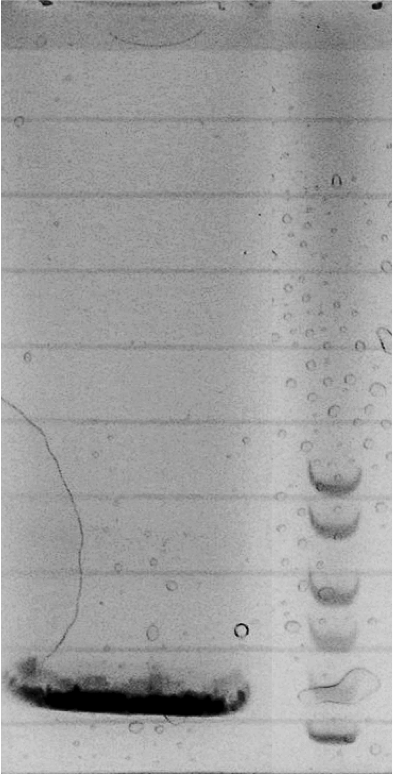
| + | Competent cells were transformed with piG0008 (F2620, ''luxR'') and selected on chloramphenicol-LB-plates. Plasmid DNA was extracted by performing a miniprep. The relevant plasmid sequence was sequenced by Microsynth using the primers oiG0001 and oiG0002. Amplification of ''luxR'' by PCR using oiG0005 and oiG0006 resulted in the fragment fiG0005 (0.8 kb). The backbone of the vector piG0040 (''lasR'' in pSEVA 181) was amplified by PCR using oiG0007 and oiG0008. This fragment, fiG0011 (3.2 kb), and fiG0005 were assembled using Gibbson assembly. Thus we formally replaced ''lasR'' by ''luxR'' to construct the ''luxR'' regulator plasmid piG0041. Competent cells were transformed with piG0041 and selected on ampicillin-LB-plates. Colony PCR using the primers oiG0035 and oiG0036 was conducted to check the size of the inserted fragment (expected bands at 1.1 and 1.2 kb). The sequence was verified by Microsynth using the same primers. | ||
| + | |||
| + | [[File:F11.png|x300px]] | ||
| + | [[File:20140712_piG0042_GA_colony_PCRs.jpg|x300px]] | ||
| + | |||
| + | |||
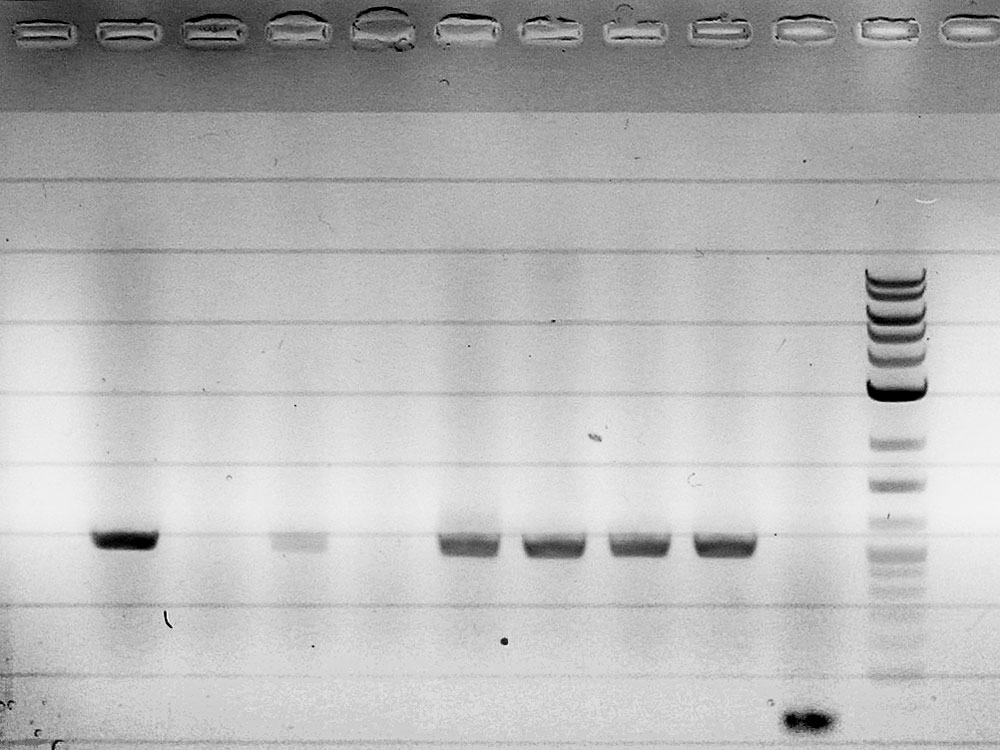
| + | ===Construction of the ''rhlR'' regulator plasmid (piG0042)=== | ||
| + | Competent cells were transformed with piG0023 (C0171, ''rhlR'') and selected on chloramphenicol-LB-plates. Plasmid DNA was extracted by performing a miniprep. The relevant plasmid sequence was sequenced by Microsynth using the primers oiG0001 and oiG0002. Amplification of ''rhlR'' by PCR using oiG0009 and oiG0010 resulted in the fragment fiG0006 (0.8 kb). The backbone of the vector piG0040 (''lasR'' in pSEVA 181) was amplified by PCR using oiG0011 and oiG0012. This fragment, fiG0012 (3.2 kb), and fiG0006 were assembled using Gibbson assembly. Thus we formally replaced ''lasR'' by ''rhlR'' to construct the ''rhlR'' regulator plasmid piG0042. Competent cells were transformed with piG0042 and selected on ampicillin-LB-plates. Colony PCR using the primers oiG0035 and oiG0036 was conducted to check the size of the inserted fragment (expected bands at 1.1 and 1.2 kb). The sequence was verified by Microsynth using the same primers. | ||
| + | |||
| + | [[File:F6.png|x300px]] | ||
| + | [[File:F12.png|x300px]] | ||
| + | [[File:20140712_piG0042_GA_colony_PCRs.jpg|x300px]] | ||
| + | |||
| + | |||
| + | ===Construction of the ''luxR'' regulator plasmids with alternative constitutive promoters (piG0046 and piG0047)=== | ||
| + | The promoter site of the ''luxR'' regulator plasmid piG0041 was mutated by QuikChange site-specific mutagenesis to produce promoters of different strength. The primers oiG0031 and oiG0032 were used to establish piG0047, a plasmid with a promoter of intermediate strength (J23111). Analoguos the primers oiG0033 and oiG0034 were used to construct piG0046, a plasmid with a weak promoter (J23109). Competent cells were transformed with piG0046 and piG0047 and selected on ampicillin-LB-plates. The sequence was verified by Microsynth using the primers oig0035 and oiG0036. | ||
| + | |||
| + | {{:Team:ETH Zurich/tpl/topbutton|green}} | ||
| + | |||
| + | {{:Team:ETH Zurich/tpl/rmbutton|green|constructionreg}} | ||
| + | |||
| + | <html> </article></html> | ||
Latest revision as of 15:11, 11 October 2014
Construction of the regulator plasmids
Thursday, July 5th
Construction of the lasR regulator plasmid (piG0040)
Competent cells were transformed with piG0028 (K553003, lasR) and selected on chloramphenicol-LB-plates. Plasmid DNA was extracted by performing a miniprep. The relevant plasmid sequence was sequenced by Microsynth using the primers oiG0001 and oiG0002. Amplification of lasR by PCR using oiG0003 and oiG0004 resulted in the fragment fiG0004 (1.0 kb). The vector piG0034 (pSEVA 181) was digested with the restriction enzymes HindIII and PacI to fiG0001 (3.0 kb). The two fragments, fiG0001 and fiG0004, were assembled using Gibbson assembly. Thus we used the plasmid backbone of piG0034 and lasR of piG0028 to construct the lasR regulator plasmid piG0040. Competent cells were transformed with piG0040 and selected on ampicillin-LB-plates. Colony PCR using the primers oiG0035 and oiG0036 was conducted to check the size of the inserted fragment (expected bands at 1.0 and 1.2 kb). The sequence was verified by Microsynth using the same primers.
Construction of the luxR regulator plasmid (piG0041)
Competent cells were transformed with piG0008 (F2620, luxR) and selected on chloramphenicol-LB-plates. Plasmid DNA was extracted by performing a miniprep. The relevant plasmid sequence was sequenced by Microsynth using the primers oiG0001 and oiG0002. Amplification of luxR by PCR using oiG0005 and oiG0006 resulted in the fragment fiG0005 (0.8 kb). The backbone of the vector piG0040 (lasR in pSEVA 181) was amplified by PCR using oiG0007 and oiG0008. This fragment, fiG0011 (3.2 kb), and fiG0005 were assembled using Gibbson assembly. Thus we formally replaced lasR by luxR to construct the luxR regulator plasmid piG0041. Competent cells were transformed with piG0041 and selected on ampicillin-LB-plates. Colony PCR using the primers oiG0035 and oiG0036 was conducted to check the size of the inserted fragment (expected bands at 1.1 and 1.2 kb). The sequence was verified by Microsynth using the same primers.
Construction of the rhlR regulator plasmid (piG0042)
Competent cells were transformed with piG0023 (C0171, rhlR) and selected on chloramphenicol-LB-plates. Plasmid DNA was extracted by performing a miniprep. The relevant plasmid sequence was sequenced by Microsynth using the primers oiG0001 and oiG0002. Amplification of rhlR by PCR using oiG0009 and oiG0010 resulted in the fragment fiG0006 (0.8 kb). The backbone of the vector piG0040 (lasR in pSEVA 181) was amplified by PCR using oiG0011 and oiG0012. This fragment, fiG0012 (3.2 kb), and fiG0006 were assembled using Gibbson assembly. Thus we formally replaced lasR by rhlR to construct the rhlR regulator plasmid piG0042. Competent cells were transformed with piG0042 and selected on ampicillin-LB-plates. Colony PCR using the primers oiG0035 and oiG0036 was conducted to check the size of the inserted fragment (expected bands at 1.1 and 1.2 kb). The sequence was verified by Microsynth using the same primers.
Construction of the luxR regulator plasmids with alternative constitutive promoters (piG0046 and piG0047)
The promoter site of the luxR regulator plasmid piG0041 was mutated by QuikChange site-specific mutagenesis to produce promoters of different strength. The primers oiG0031 and oiG0032 were used to establish piG0047, a plasmid with a promoter of intermediate strength (J23111). Analoguos the primers oiG0033 and oiG0034 were used to construct piG0046, a plasmid with a weak promoter (J23109). Competent cells were transformed with piG0046 and piG0047 and selected on ampicillin-LB-plates. The sequence was verified by Microsynth using the primers oig0035 and oiG0036.
 "
"