Team:ETH Zurich/lab/protocols
From 2014.igem.org
(→Multi site directed mutagenesis) |
(→Multi site directed mutagenesis) |
||
| Line 407: | Line 407: | ||
Method | Method | ||
| - | + | #Design mutagenesis primer(s). The targeted mutation should be in the middle of the primer. Design your primers (including the mutations) to have a Tm >=78°C. | |
| - | The targeted mutation should be in the middle of the primer | + | #Purify template plasmid from a dam+ E. coli strain via miniprep. |
| - | Design your primers (including the mutations) to have a Tm >=78°C. | + | #Set up mutagenesis PCR mix: |
| - | + | <br> | |
| - | + | ||
16 μL Water | 16 μL Water | ||
| + | <br> | ||
1.5 μL DMSO(100%) | 1.5 μL DMSO(100%) | ||
| + | <br> | ||
0.5 μL MgSO4(50 µM) | 0.5 μL MgSO4(50 µM) | ||
| + | <br> | ||
10 μL Phusion HF Buffer | 10 μL Phusion HF Buffer | ||
| + | <br> | ||
5 μL T4 Ligase Buffer | 5 μL T4 Ligase Buffer | ||
| + | <br> | ||
5 μL 10x Taq Ligase Buffer | 5 μL 10x Taq Ligase Buffer | ||
| + | <br> | ||
0.5 μL Each Mutagenesis Primer (at 40 µM) | 0.5 μL Each Mutagenesis Primer (at 40 µM) | ||
| + | <br> | ||
2 μL Reverse Biobrick Primer | 2 μL Reverse Biobrick Primer | ||
| - | 2 μL dNTPs | + | <br> |
| + | 2 μL dNTPs (100mM) | ||
| + | <br> | ||
2 μL Template | 2 μL Template | ||
| + | <br> | ||
2 μL PNK | 2 μL PNK | ||
| + | <br> | ||
2 μL Taq Ligase | 2 μL Taq Ligase | ||
| + | <br> | ||
1 μL Phusion Polymerase (hotstart) | 1 μL Phusion Polymerase (hotstart) | ||
| - | + | <br> | |
| - | 37°C for 30 minutes (this is the kinase step) | + | #Run PCR |
| - | 95°C for 3 minutes | + | *37°C for 30 minutes (this is the kinase step) |
| - | Run the following for 20 cycles: | + | *95°C for 3 minutes |
| - | 95°C for 1 minute | + | *Run the following for 20 cycles: |
| - | 55°C for 1 minute | + | **95°C for 1 minute |
| - | 65°C for 30 sec/kb of plasmid length minimum | + | **55°C for 1 minute |
| - | 4°C infinite | + | **65°C for 30 sec/kb of plasmid length minimum |
| - | + | **4°C infinite | |
| - | + | #Add 1μL DpnI restriction enzyme to the PCR tube directly. (Purification is not necessary) | |
| - | + | #Incubate 4-6 hours at 37°C. | |
| - | + | #Purify PCR product and elute into 30μL. | |
| - | + | #Transform 3μL purified DNA into highly competent cells. | |
| + | #Screen the transformants for the desired mutation using restriction digest or sequencing. | ||
| + | <br> | ||
Notes | Notes | ||
Because the inital kinase step could lead to non-specific binding, it would be best to use a hot-start polymerase for this procedure. | Because the inital kinase step could lead to non-specific binding, it would be best to use a hot-start polymerase for this procedure. | ||
Any combination of DMSO and MgSO4 has shown significant improvments in product. | Any combination of DMSO and MgSO4 has shown significant improvments in product. | ||
| + | |||
http://openwetware.org/wiki/Richard_Lab:Site_Directed_Mutagenesis | http://openwetware.org/wiki/Richard_Lab:Site_Directed_Mutagenesis | ||
Revision as of 16:00, 10 October 2014
Protocols
Materials
LB medium from dehydrated product
Suspend/dissolve the powder in 1 L of purified water:
Difco™ LB Agar, Miller – 40 g;
Difco™ LB Broth, Miller – 25 g.
Mix thoroughly.
Heat the agar medium with frequent agitation and boil for 1 minute to completely dissolve the powder.
Autoclave at 121°C for 15 minutes.
Test samples of the finished product for performance using stable, typical control cultures.
According to LB Agar, Miller; LB Broth, Miller
Preparation of antibiotics stock (1000x)
| Ampicillin (200 mg/mL) in H2O, sterile filtered |
| Kanamycin (50 mg/mL) in H2O, sterile filtered |
| Chloramphenicol (34 mg/mL) in ethanol |
| Tetracycline (10 mg/mL) in ethanol |
| Streptamycin (25 mg/mL) in H2O, sterile filtered |
SOC
| Tryptone | 20 g |
| Yeast extract | 5 g |
| NaCl | 0.5 g |
| Dissolve, then add | |
| KCl (250 mM) | 10 mL |
| MgCl2 | 5 mL |
| Autoclave, then add | |
| Sterile glucose (1 M) | 20 mL |
According to QIAGEN DNA protocols and applications
Transformation buffer 1 (TFB1)
| RbCl (100 mM) | 12.1 g |
| MnCl2·4H2O (50 mM) | 9.9 g |
| Potassium acetate (30 mM) | 2.9 g |
| CaCl2·2H2O (10 mM) | 1.4 g |
| Glycerol (15%) | 150 mL |
| Adjust pH to 5.8 with KOH | |
| Sterilize by filtration |
According to QIAGEN DNA protocols and applications
Transformation buffer 2 (TFB2)
| RbCl (10 mM) | 1.2 g |
| MOPS (10 mM) | 2.1 g |
| CaCl2·2H2O (75 mM) | 11.0 g |
| Glycerol (15%) | 150 mL |
| Adjust pH to 6.8 with KOH | |
| Sterilize by filtration |
According to QIAGEN DNA protocols and applications
Gibson Assembly reaction mixture
5x isothermal reaction buffer
| Tris-HCl pH 7.5 (1 M) | 3 mL |
| MgCl2 (2 M) | 150 μL |
| dGTP (100 mM) | 60 μL |
| dATP (100 mM) | 60 μL |
| dTTP (100 mM) | 60 μL |
| dCTP (100 mM) | 60 μL |
| DTT (1 M) | 300 mL |
| PEG-8000 | 1.5 g |
| NAD (100 mM) | 300 μL |
Aliquots can be stored at -20 °C
Assembly master mixture
| Isothermal reactio buffer (5x) | 320 μL |
| T5 exonuclease (10 U/μL) | 0.64 μL |
| Phusion DNA polymerase (2 U/μL) | 20 μL |
| Taq DNA ligase (40 U/μL) | 160 μL |
| H2O | 1.2 mL |
Aliquots of 15 μL can be stored at -20 °C
Methods
Preparation of competent E. coli
Day before: Preparation of a Top10 cells preculture in LB-medium containing streptomycin (25 μg/mL)
- Addition of 1 mL overnight preculture to 100 mL LB-medium + streptomycin (25 μg/mL)
- Cultivate culture at 37 °C, 220 rpm until it reaches an OD600 of 0.5
- Cool culture for 5 min on ice and centrifuge it for 5 min at 4 °C, 4000 g
- Discard the supernatant and resuspend the cells in cold TFB1 buffer (30 mL, 4 °C)
- Keep the suspension on ice for 90 min
- Centrifuge the suspension for 5 min at 4 °C, 4000 g and discard the supernatant
- Resuspend the cells in 4 mL cold TFB2 buffer
- Make aliquots of 100 μL and freeze the aliquots in dry ice in ethanol
- Store aliquots at -80 °C
According to QIAGEN DNA protocols and applications
Preparation of DNA from iGEM kit
- Add 10 μL H2O to appropriate well, wait for 5 min, transfer into sterile tube
- Use 2 μL to transform competent cells
Biological parts used from iGEM kit:
| Part-ID | Function |
| BBa_F2620 | tetR-luxR-pLuxR |
| BBa_K084007 | pLacI-lasI |
| BBa_C0070 | rhlI-LVA |
| BBa_C0171 | rhlR |
| BBa_J23100 | constitutive promoter |
| BBa_K553003 | promoter-RBS-lasR-Term |
| BBa_C0161 | luxI |
For confirmation all plasmids used from the iGEM kit were sequenced
Transformation of competent E. coli
- Thaw the competent cells on ice
- Add 1 μL DNA (0.2 - 200 ng) to 50-100 μL competent cells
- Leave sample on ice for approximately 20 min
- Heat shock the cells for 90 s at 42 °C
- Add 500 μL of SOC to the sample
- Let the cells recover for 60 min at 37 °C, 220 rpm
- Plate appropriate amount of cell suspension (50 - 200 μL) on LB-agar-plates containing the appropriate antibiotic
- Let bacteria grow overnight at 37 °C
According to QIAGEN DNA protocols and applications
Plasmid preparation
Day before: Preparation of a preculture in LB-medium containing the appropriate antibiotics. The amount of cell culture required for plasmid preparation depends on the copy number of the plasmid (between 5 and 20 mL)
- Centrifuge the preculture in appropriate tubes for 10 min at 4000 g
- Carefully remove the supernatant
- Resuspend pellet in 200 μL resuspension solution
- Add 200 μL lysis solution and invert the tube gently 1 to 2 times
- Lysis should not exceed 5 min
- Add 350 μL neutralization solution and invert the tube 4 to 6 times
- Centrifuge the suspension for 10 min at 12 000 rcf
- Prepare the columns by adding 500 μL of column preparation solution and centrifuging it for 1 min at 12 000 rcf. Discard the flow-through
- Transfer the supernatant of the centrifuged samples onto columns
- Spin for 1 min at 12 000 rcf and subsequently discard the flow-through
- Add 750 μL wash solution and spin the loaded column for 1 min at 12 000 rcf. Discard the flow-through
- Dry the columns by centrifuging them for 1 min at 12 000 rcf
- Place columns in new collection tubes
- Elute the plasmid DNA with 50 μL ddH2O to increase the plasmid concentration
The Sigma-Aldrich GenElute™ Plasmid Miniprep Kit was used
Preparation of samples for sequencing at Microsynth
Add 12 μL DNA (60-100 ng/μL) to 3 μL of the corresponding primer (10 μM).
PCR procol for phusion DNA polymerase
| Components | 20 μL total reaction volume | 50 μL total reaction volume |
|---|---|---|
| 5x Phusion HF buffer | 4 μL | 10 μL |
| 10 mM dNTPs | 2 μL | 5 μL |
| Forward Primer (10 μM) | 1 μL | 2.5 μL |
| Reverse Primer (10 μM) | 1 μL | 2.5 μL |
| DMSO | 0.6 μL | 1.5 μL |
| Phusion DNA polymerase | 0.2 μL | 0.5 μL |
| DNA | 40-200 ng | 40-200 ng |
| H2O | add to reach a total volume of 20 μL | add to reach a total volume of 50 μL |
Restriction Endonuclease Reaction (double digestion)
| 1-2.5 μL restriction endonuclease 1 |
| 1-2.5 μL restriction endonuclease 2 |
| 5 μL Cut Smart Buffer |
| 1-3 μg template DNA |
| add H2O to reach a total volume of 50 μL |
Enzymes, buffers and protocol are from New England BioLabs
Dephosphorylation of 5’-ends of DNA using CIP
Add 1 unit of CIP for every 1 pmol of DNA ends (about 1 μg of a 3 kb plasmid) and incubate at 37 °C for 30–60 min.
Enzymes, buffers and protocol are from New England BioLabs
DNA Purification by Centrifugation
A. Dissolving the Gel Slice
Following electrophoresis, excise DNA band from gel and place gel slice in a 1.5 ml microcentrifuge tube. Add 10 µl Membrane Binding Solution per 10 mg of gel slice. Vortex and incubate at 50–65 °C until gel slice is completely dissolved.
or
B. Processing PCR Amplifications
Add an equal volume of Membrane Binding Solution to the PCR mix.
- Insert SV Minicolumn into Collection Tube.
- Transfer dissolved gel mixture or prepared PCR product to the Minicolumn assembly. Incubate at room temperature for 1 min.
- Centrifuge at 16 000 rcf for 1 min. Discard flowthrough and reinsert Minicolumn into Collection Tube.
- Add 700 µL Membrane Wash Solution (ethanol added). Centrifuge at 16 000 rcf for 1 min. Discard flowthrough and reinsert Minicolumn into Collection Tube.
- Repeat the step before with 500 µL Membrane Wash Solution. Centrifuge at 16 000 rcf for 5 min.
- Empty the Collection Tube and recentrifuge the column assembly for 1 min with the microcentrifuge lid off to allow evaporation of any residual ethanol.
- Carefully transfer Minicolumn to a clean 1.5 mL microcentrifuge tube.
- Add 50 µL of Nuclease-Free Water to the Minicolumn. Incubate at room temperature for 1 min. Centrifuge at 16 000 rcf for 1 min.
- Discard Minicolumn and store DNA at 4 °C or –20 °C.
According to Promega Wizard SV Gel and PCR Clean-Up System, Quick protocol
Agarose gel electrophoresis
- For a 5 cm x 6 cm gel add 0.25 g Agarose to 25 mL TAE (0.5x) and heat up in microwave to dissolve it
- Let the solution cool down to approximately 50 °C
- Add nucleic acid dye (e.g. 2.5 μL peqGREEN) and mix
- Pour the solution in a tray using an appropriate comb
- Add loading dye (e.g. NEB purple loading dye) to the samples
- Fill the samples and ladder in the wells
- Run the gel at 135 V
Preparation of cryostocks
- Cultivate bacteria (37 °C) in medium with antibiotic (e.g. 5 mL LB, 5 μL amp + 500 μL preculture) until they are in log phase (OD=0.8-1.2)
- Add 750 μL sterile glycerol (30%) to 750 μL bacteria culture in a screw top tube
- Freeze the glycerol stock tube at -80 °C
Site-directed mutagenesis
| 14 μL H2O |
| 2 μL HF buffer |
| 1.6 μL dNTPs |
| 0.5 μL of primer 1 |
| 0.5 μL of primer 2 |
| 0.4 μL Phusion polymerase |
| 1 μL template DNA (2-20 ng) |
- Run PCR
- Digestion of template DNA: Addition of 1 μL DpnI, 1h at 37 °C
- Heat inactivation of DpnI: 20 min at 80 °C
Protocol based on QuikChange Site-Directed Mutagenesis
Gibson Assembly
One-step isothermal DNA assembly protocol: the exonuclease amount is ideal for the assembly of DNA molecules with 20–150 bp overlaps
- Mix the backbone and PCR fragments in 5 µL total volume in equimolar amounts
- Thaw the Gibson assembly reaction mixture on ice
- Add DNA mixture (5 µL) to the reaction mixture (15 µL)
- Run the reaction for 30-60 min at 50 °C
- Subsequently the reaction mixture (5 uL are enough) can be used directly to transform competent cells (75 uL)
Multi site directed mutagenesis
Single Primer Method
This protocol only uses one oligo per mutation site (cuts the oligo cost in half) and can mutate more than one site at a time. While this protocol is less reliable than the previous protocol (only ~25% of colonies will contain the desired mutation(s)), if you're doing multiple mutations on the same piece of DNA than this can save you many days of work by just sequencing a few more colonies. In this protocol there is an initial Kinase (phosphorylation) reaction to allow your primers to be available for ligation. The polymerase will extend from one primer to another and then the Taq ligase will seal the nick. This allows multiple mutations to be done at the same time.
Method
- Design mutagenesis primer(s). The targeted mutation should be in the middle of the primer. Design your primers (including the mutations) to have a Tm >=78°C.
- Purify template plasmid from a dam+ E. coli strain via miniprep.
- Set up mutagenesis PCR mix:
16 μL Water
1.5 μL DMSO(100%)
0.5 μL MgSO4(50 µM)
10 μL Phusion HF Buffer
5 μL T4 Ligase Buffer
5 μL 10x Taq Ligase Buffer
0.5 μL Each Mutagenesis Primer (at 40 µM)
2 μL Reverse Biobrick Primer
2 μL dNTPs (100mM)
2 μL Template
2 μL PNK
2 μL Taq Ligase
1 μL Phusion Polymerase (hotstart)
- Run PCR
- 37°C for 30 minutes (this is the kinase step)
- 95°C for 3 minutes
- Run the following for 20 cycles:
- 95°C for 1 minute
- 55°C for 1 minute
- 65°C for 30 sec/kb of plasmid length minimum
- 4°C infinite
- Add 1μL DpnI restriction enzyme to the PCR tube directly. (Purification is not necessary)
- Incubate 4-6 hours at 37°C.
- Purify PCR product and elute into 30μL.
- Transform 3μL purified DNA into highly competent cells.
- Screen the transformants for the desired mutation using restriction digest or sequencing.
Notes
Because the inital kinase step could lead to non-specific binding, it would be best to use a hot-start polymerase for this procedure.
Any combination of DMSO and MgSO4 has shown significant improvments in product.
http://openwetware.org/wiki/Richard_Lab:Site_Directed_Mutagenesis
Preparation of alginate beads
- Harvest bacteria in exponential phase (OD between 1.5 and 2.0)
- Centrifuge the culture for 10 min at 4000 rcf
- Resuspend and dilute the pellet in sterile NaCl solution (0.9%)
- Add bacteria-NaCl-suspension to sterile alginate (2.5%) so as to reach an alginate concentration of 2%
- Fill the suspension into a sterile syringe
blablabla
1 bead: approximatly 14.5 µl alginate-NaCl
Devices
NanoDrop 2000, UV-Vis Spectrophotometer
A Thermo Scientific NanoDrop® 2000 spectrophotometer was used to determine the concentrations of our plasmids after miniprep.
This microvolume spectrophotometer can be used to measure the concentration and purity of DNA, RNA or protein samples. Having a wide spectral range (190-840nm) allows for measuring a variety of samples types such as peptides, DNA, RNA, purified protein and even gold nanoparticles. Even for highly concentrated samples no dilutions are required. Results are obtained in less than 15 seconds from sample pipetting to wiping the pedestal clean. For the measurement only 0.5 – 2uL of the samples are needed. The software with an intuitive user interface allows for classical NanoDrop applications such as nucleic acids, protein A280, colorimetric assays and new user defined custom methods.
Please visit the NanoDrop webpage for more information.
Tecan Infinite M200 Pro™
For performing fluorescence measurements we used a Tecan Infinite M200 ProTM equipped with the Tecan iControl TM software.
The Infinite 200 PRO is a user-friendly and affordable multimode reader, designed to cater for the needs of today’s applications. Based on the highly successful Infinite 200 series of microplate reader, the Infinite 200 PRO can provide a full range of leading detection methods in one easy-to-use modular instrument, with either Quad4 monochromator™ or filter-based technologies. Users can select the desired modules to create the perfect reader for their needs, with the option to upgrade as requirements change. The Infinite 200 PRO offers excellent sensitivity, multiplexing capabilities and high format flexibility, including 6- to 384-well microplates, PCR plates, cuvettes and Tecan’s patentedNanoQuant Plate™ for low sample volumes.
- For uncompromised performance in all detection modes, the Infinite 200 PRO uses advanced optics and high-performance detectors, optimized for the requirements of fluorescence, luminescence and absorbance reading. The Infinite 200 PRO offers a wide range of detection modes, including:
- Fluorescence intensity (top and bottom reading) (UV – NIR & wavelength scanning)
- Fluorescence resonance energy transfer (FRET)
- Time resolved fluorescence energy transfer (TR-FRET) like HTRF®
- Time resolved fluorescence (TRF)
- Fluorescence polarization (FP)
- Flash luminescence
- Glow luminescence
- Dual-color luminescence (including BRET 1 & BRET 2 applications)
- Absorbance (UV – NIR & wavelength scanning)
- AlphaScreen® and AlphaLISA® technology
- Injectors system for up to two reagents
- Temperature control
- Small sample measurement in NanoQuant Plate
Please visit the Tecan webpage for more information.
Tecan iControl™ software
Our Tecan is equipped with i-control, the easy-to-use microplate reader software. It allows the user to define the workflow for each application. Each workflow is easily created by dragging and dropping the processing steps into a sequence according to the assay protocol. The application workflow is then visible to the user and can be saved for future use. Data are easily managed and exported to Windows® compatible formats (Excel®). Intuitive Easy set up of workflows via drag & drop Workflows are visible to the user Workflows may be saved as measurement scripts On-line data presentation in Excel® Graphical definition of measurement range Plate definition editor Enhanced data management Export of data into user-defined Excel® templates Flexible User sets up the workflow to suit his own application Multilabel measurements for multiplexed assays Different kinds of kinetic measurements Supports ratio mode also for well kinetic measurements
Please visit the Tecan software webpage for more information.
Novaspec Plus™ Visible Spectrophotometer
Novaspec™ Plus Visible Spectrophotometer is ideal for general biotech laboratory use. There are stored methods for the standard colorimetric methods of Bradford, BCA, Biuret and Lowry in addition to the basic modes of absorbance, transmittance,OD600 and concentration measurements. The use of diode array technology permits rapid wavelength scans to be made as well and, as there are no moving parts, make for a highly reliable and low maintenance product (Fig 1). The large backlit display enables the graphical results for wavelength scans, kinetics assays (including slope calculation for rate/activity studies) and standard curves to be viewed (Fig 2).Up to 99 methods for such experiments can be stored for instant recall and use by a laboratory assistant. Novaspec Plusis delivered with Grafico, a PC utility software package, and the requisite serial lead, providing the user with the means to capture, print and store data from the instrument (with a time/date stamp) onto a PC so that a results log can be built up or data exported to Microsoft™ Excel.
The cell holder supplied with Novaspec Plus accepts standard 10-mm pathlength glass or plastic cells (adapters are availableto convert it to accept 10-, 12- and 16-mm diameter test tubes).It can be removed for cleaning, or flushed through with water in situ, if spillages occur. For users who require thermostatting, particularly for kineticsstudies and use with test kits, a special version of Novaspec Plus is available with a factory fitted heated cell holder (37 °C only);note that this cell holder cannot be added retrospectively.
Features:
- “Flash Scan” diode array
- Stored protein methods
- Bacterial cell culture measurement at OD600
- Kinetics for activity studies
- Stored methods
Please visit the Novaspec™ Plus Visible Spectrophotometer webpage for more information.
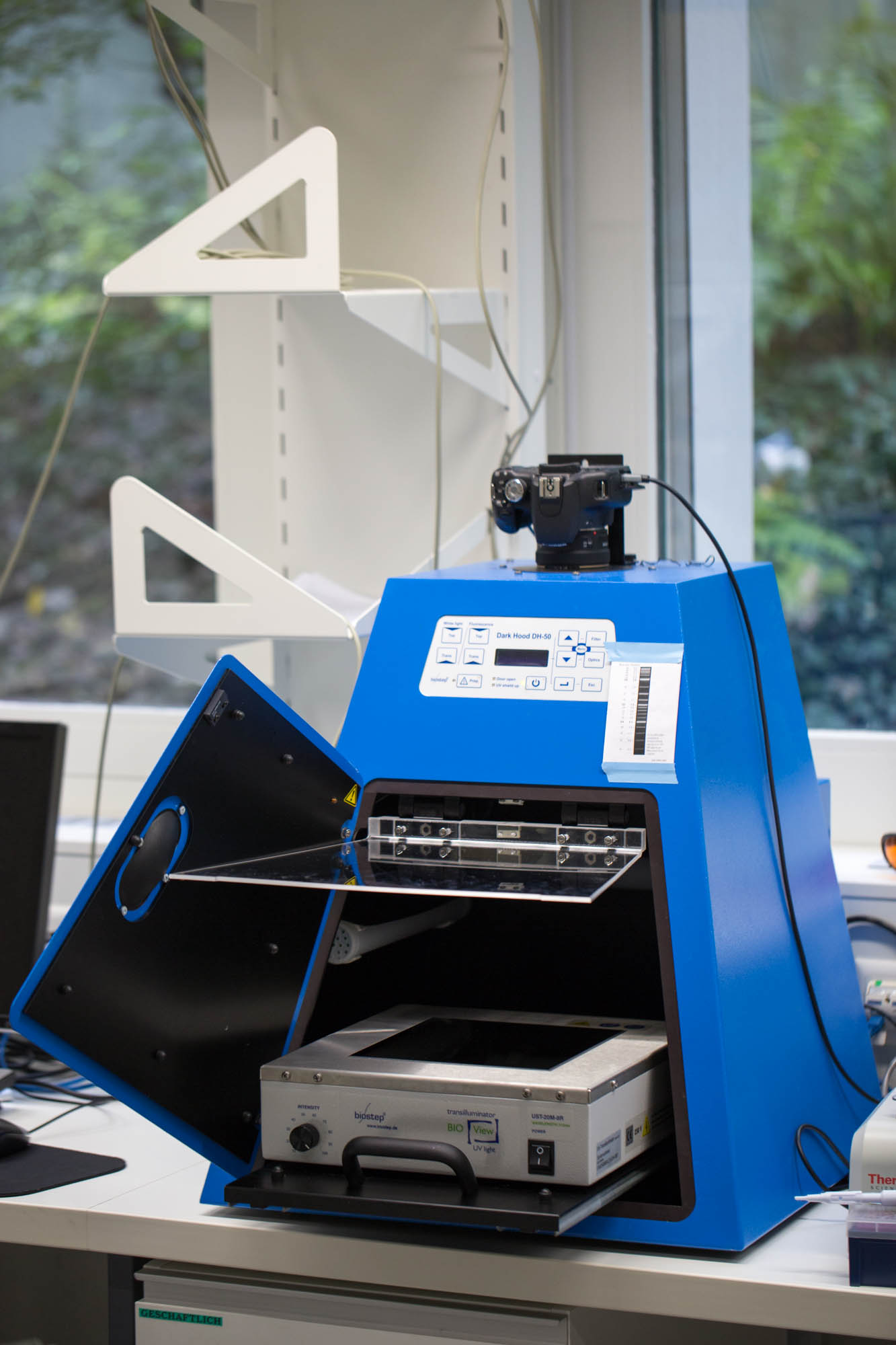
Biostep Dark-Hood DH-50™ and the Argus-X1™ software
Pictures of our gels, long term imaging of our chips with beads containing GFP-expressing cells and pictures of our plates with bacterial colonies were recorded with the DH-50™ and the Argus-X1™ software.
Dark Hood DH-50 with control panel, display, sliding table, UV protection, for CCD camera stationary dark hood with large door sliding table to put on a transilluminator incl. UV protection shield – no additional protection necessary dimmable white top-light by LEDs automatic UV cut-off when opening the UV protection shield preparative function enables observation of gels under UV light control panel with keypad and LCD display
Functionality: transilluminator: on/off white top-light: on/off, dim function preparative function optionally available: control of zoom, aperture and focus by motor zoom objective optionally available: storage of application methods repeatable
- transilluminators with dimensions (W x D x H): 32.5x32.2x10.5cm; corresponds to the series of biostep UST, USDT, UXFT, BST, GST, YST, RST, WST
optionally upgradeable with UV protection shield, filter wheel, white transmission light, UV or epi light in the visible range
Please visit the Biostep Dark-Hood DH-50™ webpage for more information.
Biostep argusX1™ basic licence
PC-software for controlling the camera basic modules Felix 1000/2000, 5000/6000/7000 and the transmission scanners of the series ViewPix 900/1300
Attention: Imperatively necessary: module for controlling the camera or scanner!!!
General software functions: integrated database for administration of the images copying and cropping of single images or group images within a database or even between database to database if the additional module Project administration exists deletion of single images or group images functions for image processing incl. Undo rotating in arbitrary angles and around 90°, 180° reflecting in horizontal and vertical direction inverting cropping various filter functions marking of the images with text, rectangle and line automated generation and numeration of sample names after the acquisition memo function: input of a note for each image and printout of the same on a protocol history for documentation of the acquisition parameters for each image overexposure control data export into other Windows applications data import of images in different formats creation of individual print reports quick print selection of the export format, export path, print report for quick print selection of the user language: German, English, Spanish, French or Italian control of the functions a of biostep transilluminators with PC interface
Please visit the Biostep argusX1™ basic licence webpage for more information.
Eppendorf Centrifuge 5810 R
Centrifuge 5810/5810 R is a workhorse for medium to high-throughput laboratories. It combines extraordinary versatility and capacity for both tubes and plates with an extraordinary compact footprint. With the new rotor generation the capacity of Centrifuge 5810/ 5810 R increases to max. 4 x 750 mL (or max. 28 x 50 mL / 56 x 15 mL). Considering its small footprint, Centrifuge 5810/5810 R is the most compact 3 liter centrifuge currently available in the market.
Features: Swing-bucket rotors and adapters accommodate tubes and bottles from 0.2 mL to 750 mL Plate rotors for centrifugation of all types of MTP, PCR, cell culture, or deepwell plates Fixed-angle rotors for high-speed molecular biology applications in tubes from 0.2 mL to 85 mL High centrifugation speed of up to 20,913 x g (14,000 rpm) Aerosol-tight Eppendorf QuickLock® caps and lids for easy, one-hand operation Centrifuge lid with soft-touch lid closure Low access height of 29 cm for easy loading and unloading of samples Quiet operation to improve your work environment Compact footprint saves valuable bench space Automatic rotor recognition and imbalance detection for maximum operational safety Additional features of refrigerated Centrifuge 5810 R: Temperature range from -9 °C to 40 °C FastTemp function for fast pre-cooling Standby cooling function holds temperature when centrifuge is not in use ECO shut-off engages after 8 hours of non-use to reduce energy consumption and to extend compressor life Dynamic compressor control (DCC) technology for optimized cooling performance
Please visit the Eppendorf Centrifuge 5810 R webpage for more information.
Eppendorf Centrifuge 5424 R
This highly advanced 24-place model is unbelievable quiet and equipped with a number of innovative features. The Centrifuge 5424 and Centrifuge 5424 R set new standards for silence, speed and simplicity – for others to follow! The micro centrifuge standard in your lab – refrigerated. It features all the benefits already combined in the Centrifuge 5424 and adds the patented cooling feature for temperature sensitive samples.
Product features of Centrifuge 5424 and 5424 R Very compact size Very silent operation, even without the rotor lid Low access height – for easy loading and unloading of the centrifuge Can be equipped with four different rotors: - Standard 24-place rotor, aerosol-tight - 24-place rotor, aerosol-tight, PTFE coated for increased chemical resistance - 18-place Kit rotor for processing spin-columns with open tube lids - 4 × 8 PCR strip rotor Two different operation panel Rotary knobs for fast and easy setting of parameters Foil version for easier cleaning of the front panel Automatic lid opening following centrifuge run – prevents sample warming (can be optionally deactivated (only 5424)) Separate Short-spin button – for fast and short spins SOFT-brake to protect delicate samples
Additional cooling features of refrigerated Centrifuge 5424 R
Patented compressor technology minimizes vibrations – protects your precious sample!
Temperature range: –9 °C to +40 °C
Small footprint and low access height of 26 cm, as refrigeration unit is at the rear of the unit
Maintains constant 4 °C at max. speed
Fast Temp for rapid pre-cooling of the centrifuge within 8 minutes from ambient temperature 23 °C
Continuous cooling maintains temperature when centrifugation process is not active
Please visit the Eppendorf Centrifuge 5424 R webpage for more information.
Harvard Pump 11 Plus
Our team used the syringe pump for fabricating the alginate beads containing our bacteria.
Harvard Apparatus’ Syringe Pump 11 Plus combines smoother flow and updated features to create a high performance syringe pump with a basic syringe pump price!
Bright Display and Easy-To-Use Interface A new, two-line 16 character vacuum fluorescent display and six membrane keys make this a powerful, easy-to-use syringe pump. Only two entries are required to start pumping; Syringe ID and flow rate. The flow rate can be changed while the pump is running.
Two Modes of Operation, Constant Flow Rate & Volume Dispense The new Pump 11 Plus will operate continuously in RATE mode or accurately dispense a specific amount of fluid in VOLUME mode.
Versatile Select from two different models. The standard Pump 11 Plus model has the following features: • Infusion Only • End of Travel Limit Stop • Anti-Siphon Bracket
Non-Volatile Memory New micro-stepping pump profiles deliver very smooth and consistent flow over the entire flow rate range. These pumps now feature non-volatile memory. The pump remembers its last syringe size, flow rate used and configuration settings. An advanced universal power supply means there is no need to change AC line switches, fuses or wires.
Please visit the Harvard Pump 11 Plus webpage for more information.
BD LSRFortessa™ Flow Cytometer System
In order to determine the exact sensivity shift and expression level of GFP with the mutated promoters the BD LSRFortessa™ Flow Cytometer System was used for the measurement,FACSDiva™ for the recording of the data and the FlowJo™ software for the data analysis.
BD LSRFortessa™ Flow Cytometer System
The BD LSRFortessa™ cell analyzer offers the ultimate in choice for flow cytometry, providing power, performance, and consistency. Designed to be affordable and expandable, the BD LSRFortessa has the flexibility to support the expanding needs of multicolor flow cytometry assays.The BD LSRFortessa analyzer can be configured with up to 4 lasers— blue, red, violet, and UV—which enables the detection of up to 18 colors simultaneously. The BD LSRFortessa may be upgraded subsequently with additional or new lasers from BD, as future requirements dictate.Innovative Designs for Optical Efficiency Built on a proven platform, BD LSR analyzers feature innovative designs for both the excitation optics and collection optics that reduce excitation losses and improve light collection efficiency. The result is optical efficiency that delivers maximum sensitivity and resolution for multicolor applications by reducing excitation laser light loss, and improved fluorescent signal collection.Special Order Research ProgramFor research needs at the leading edge of biomedical discovery, the BD LSRFortessa special order research program offers the latest innovations in laser technology to researchers to enable future upgrades with new excitation, wavelength, and power choices. The program lets customers fully customize configurations to deliver added flexibility and power to support advanced research.
Please visit the BD LSRFortessa™ Flow Cytometer System webpage for more information.
FACSDiva™ software
BD FACSDiva™ software is a collection of rich tools for flow cytometer and application setup, data acquisition, and data analysis that help streamline flow cytometry workflows for today’s busy laboratory.
BD FACSDiva software provides new features to help users integrate flow systems into new application areas, including index sorting for stem cell and single-cell applications, as well as automation protocols for high-throughput and robotic laboratories.
Please visit the FACSDiva™ software webpage for more information.
 "
"