Team:ETH Zurich/achievements
From 2014.igem.org
(→Modeling) |
|||
| Line 43: | Line 43: | ||
== Modeling == | == Modeling == | ||
=== [https://2014.igem.org/Team:ETH_Zurich/modeling/int Integrase theoretical characterization] === | === [https://2014.igem.org/Team:ETH_Zurich/modeling/int Integrase theoretical characterization] === | ||
| + | [[File:ETH_Zurich_Integrases_sites.png|300px|left|link=https://2014.igem.org/Team:ETH_Zurich/modeling/int]] | ||
Based on the data presented in Bonnet et al.'s publication "Amplifying genetic logic gates"<sup>[[Team:ETH_Zurich/project/references#refEmergence|[9]]]</sup>, we parameterized a detailed model, resulting in predictions for a significant number of parameters currently not documented in the literature, such as binding affinity of integrases to their sites on the DNA. | Based on the data presented in Bonnet et al.'s publication "Amplifying genetic logic gates"<sup>[[Team:ETH_Zurich/project/references#refEmergence|[9]]]</sup>, we parameterized a detailed model, resulting in predictions for a significant number of parameters currently not documented in the literature, such as binding affinity of integrases to their sites on the DNA. | ||
| - | |||
| + | |||
| + | <br clear="all" /> | ||
=== [https://2014.igem.org/Team:ETH_Zurich/modeling/overview Predicting the whole system's behavior with a comprehensive fully derived model] === | === [https://2014.igem.org/Team:ETH_Zurich/modeling/overview Predicting the whole system's behavior with a comprehensive fully derived model] === | ||
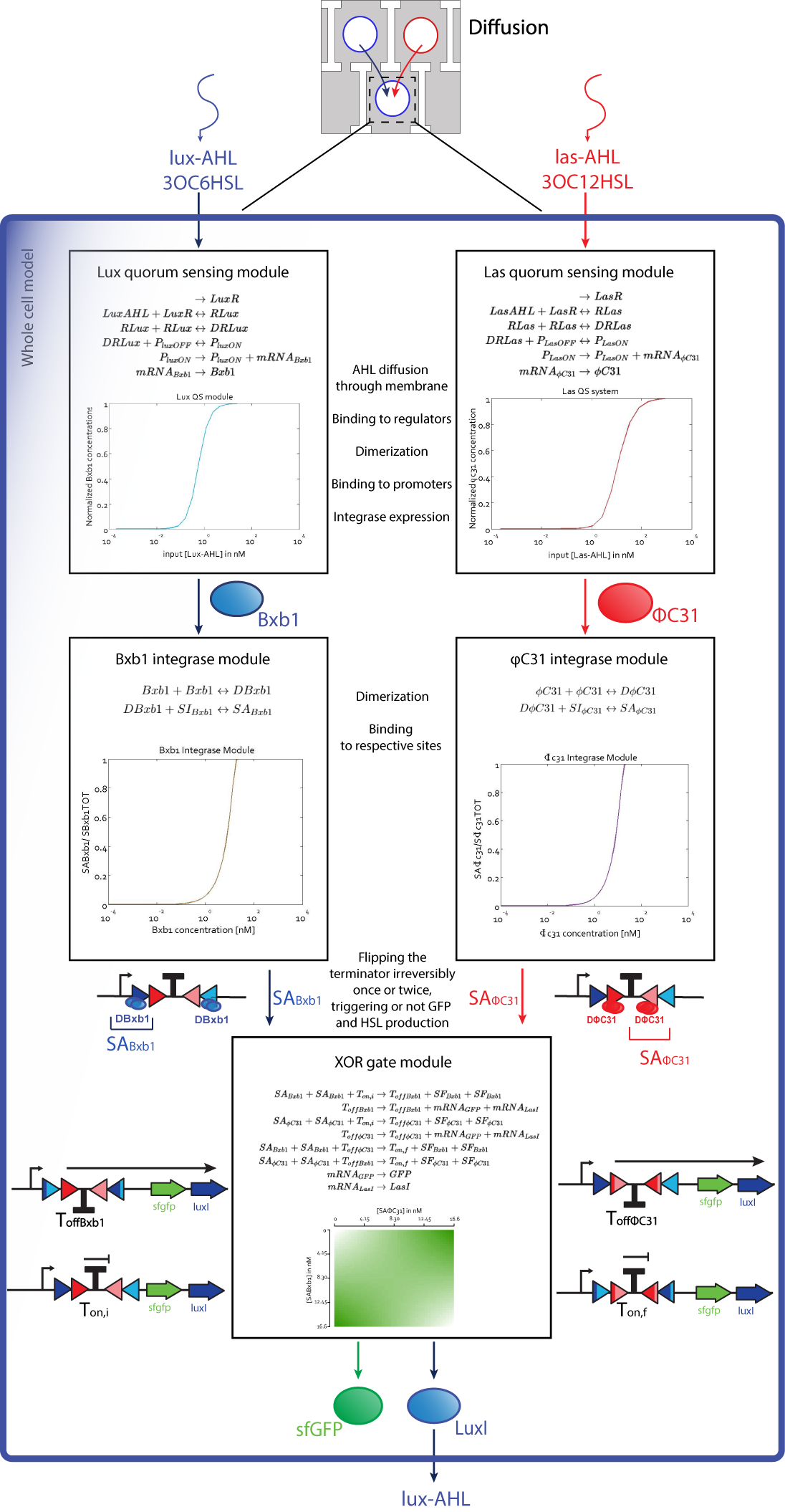
We have a thorough model for the whole Mosai''coli'' system based on a detailed study of every module of this system, including quorum sensing, integrases, and diffusion. We derived the formulae from mass action kinetics and state precisely which approximations we do and why we think we are justified to do so. All our steady state simulations and some of our dynamic simulation results fit to experimental data from our own experiments or found in literature. | We have a thorough model for the whole Mosai''coli'' system based on a detailed study of every module of this system, including quorum sensing, integrases, and diffusion. We derived the formulae from mass action kinetics and state precisely which approximations we do and why we think we are justified to do so. All our steady state simulations and some of our dynamic simulation results fit to experimental data from our own experiments or found in literature. | ||
Revision as of 01:36, 18 October 2014
Achievements
We have achieved many of the goals we planned at the beginning of our project. These achievements are well distributed between experimental results, theoretical work and human reflection and socialisation. As a team of seven students, we all contributed to all achievements and this is why everyone of us can feel the fruitfulness of such a wide project.
Lab
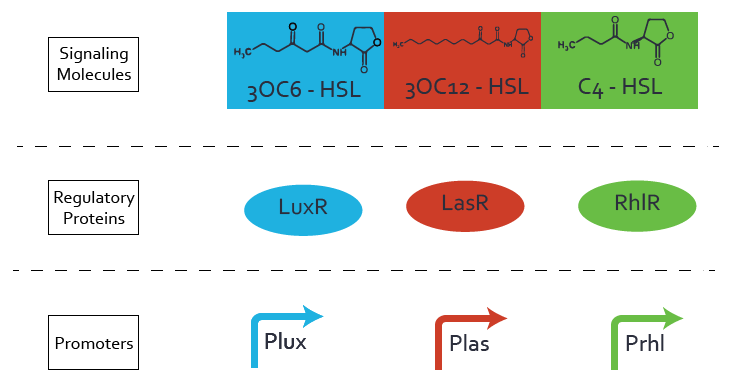
Quorum sensing crosstalk full characterization
We highlighted 3 levels of crosstalk between Las, Lux and Rhl quorum sensing systems and fully quantitatively characterized all possible combinations of crosstalk at these three levels.
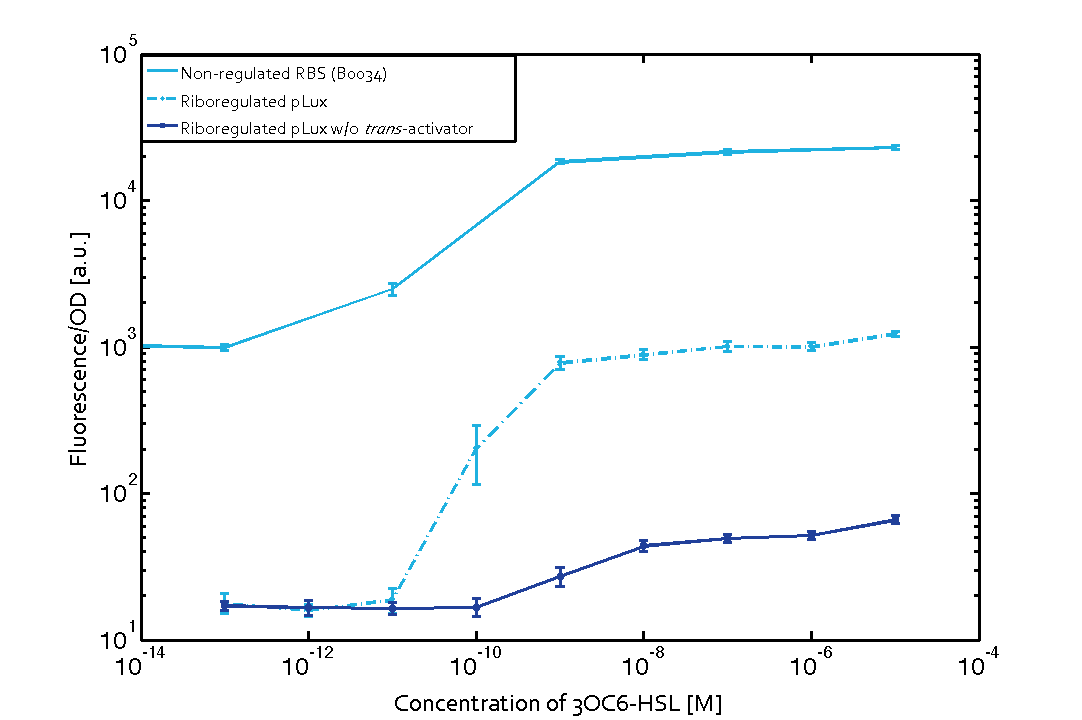
Characterizing and reducing leakiness in quorum sensing systems
We quantitatively characterized leakiness of the three quorum sensing systems (Las, Lux and Rhl). We have designed a new BioBrick for reducing leakiness of these systems and demonstrated its efficiency for the lux system, which is the most leaky of the three. We improved the signal over noise ratio for this promoter by a factor of 6 while reducing the basal expression by a factor of 60.
Novel millifluidic chip construction method
We designed a rapid prototyping approach for creating millifluidic chips: we use a 3D-printing device to create the mold for PDMS.
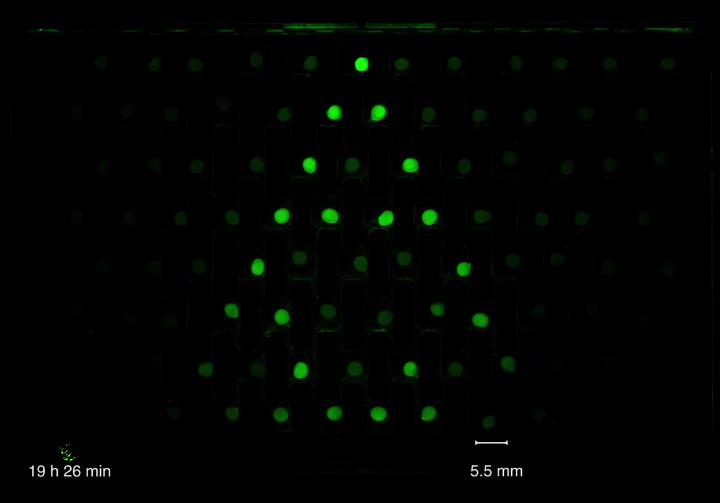
Bead to bead communication
We accomplished to produce alginate encapsulated E. coli culture beads. The cells within the beads could survive during the time of our assays and still produce green fluorescent proteins.Bead to bead communication could be observed by placing a sender bead, containing AHL producing cells, into one well of our millifluidic chip connected to another well containing a receiver bead, which contains cells, that produce green fluorescent proteins upon AHL induction.
This signal transfer could also be observed using cells, that can produce AHL and be induced by AHL in order to produce a fluorescent reporter as well as the signal itself again. Placed on a grid in our millifluidic chip, signal propagation from one single induced bead over the whole chip was could be achieved.
Modeling
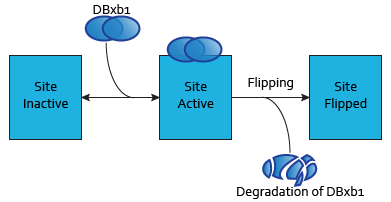
Integrase theoretical characterization
Based on the data presented in Bonnet et al.'s publication "Amplifying genetic logic gates"[9], we parameterized a detailed model, resulting in predictions for a significant number of parameters currently not documented in the literature, such as binding affinity of integrases to their sites on the DNA.
Predicting the whole system's behavior with a comprehensive fully derived model
We have a thorough model for the whole Mosaicoli system based on a detailed study of every module of this system, including quorum sensing, integrases, and diffusion. We derived the formulae from mass action kinetics and state precisely which approximations we do and why we think we are justified to do so. All our steady state simulations and some of our dynamic simulation results fit to experimental data from our own experiments or found in literature.
Human Practice
Detailed investigation on the question of complexity through seven interviews of experts, a survey and our own reflections
We conducted a deep study of complexity, how it emerges, and how we should deal with it. By interviewing experts from different fields and by listening to the public with a survey, we have broaden our views. Our own biological project also raised questions on this subject and we finally integrated everything that we learnt in a reflection of our own.
Bringing synthetic biology to the public
Last but not least, we have been communicating our enthusiasm for the field of synthetic biology during the whole project, through school visits, open house days, science slams, social media... We are commited to make our project, and synthetic biology in general, understandable and interesting for everyone. This is also why we placed some "simplify" buttons all over our wiki, each time a scientific text could be hermetic to the public.
Collaborations
Successful collaboration with Colombia, Helsinki and Edinburgh iGEM teams
We collaborated with iGEM teams from Colombia and Helsinki by responding to Colombia team's challenge to find out low budget protocols. Our cheap PCR technique has been as successful as a standard PCR.
We theoretically fused our project with Edinburgh's iGEM team's project on metabolic wiring. This fusion gave us a promising alternative solution to the crosstalk issues that arise when using quorum sensing only.

| 
| 
|
 "
"