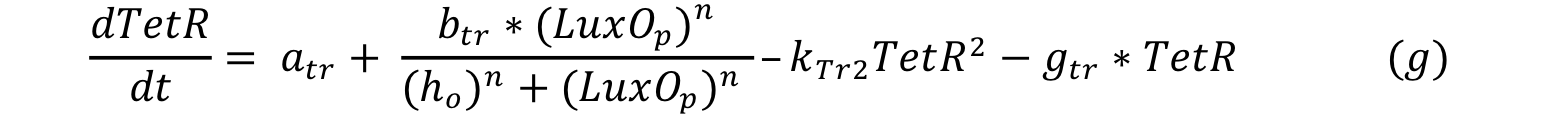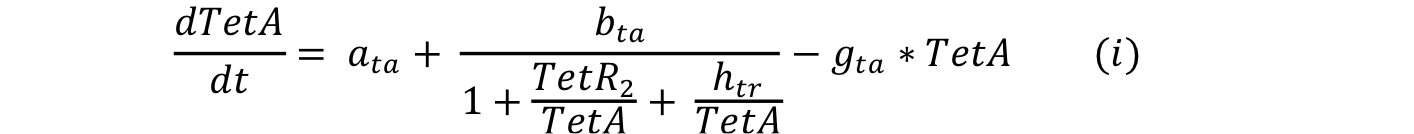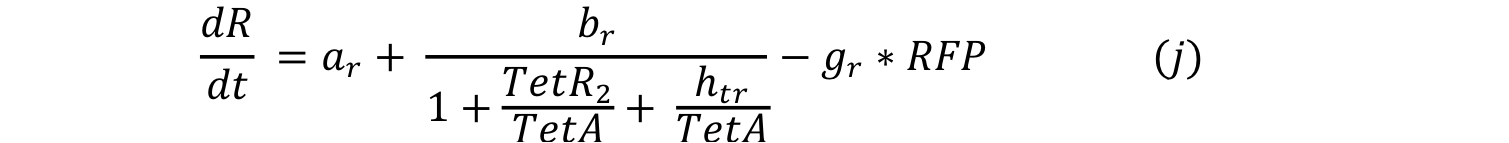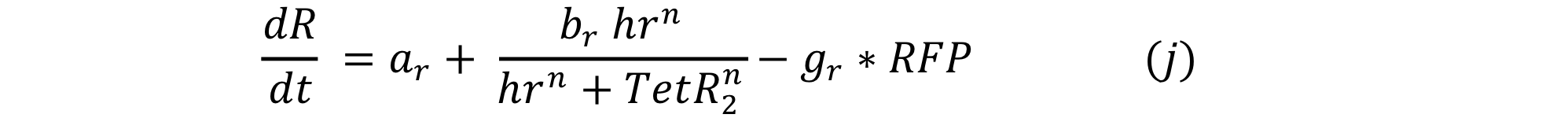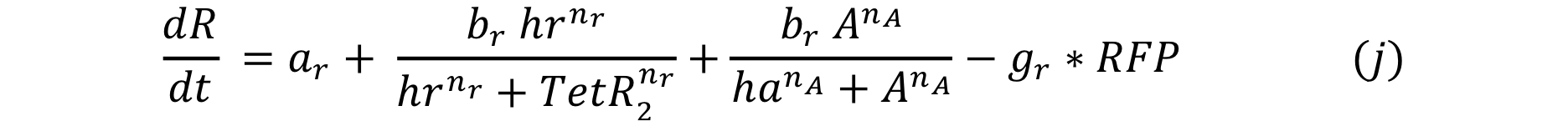Team:Colombia/Deterministic
From 2014.igem.org
Deterministic model
Finding the mean behaviour of the system...
The first step is to develop a deterministic model based in differential equations. With the ordinary differential equations we can describe quantitatively the system at molecular level.
Using the law of mass action is possible to model most of the metabolic networks. For each substance of the system, we express the changes of concentration trough time. We consider processes related with the export and import of a molecule into the cell, the production by a gene or a chemical reaction. All these terms depend on reaction kinetics that could be expressed as a simple multiplication or a complex expression like a Hill equation.
Signal transduction →
The receptor CqsS acts as a kinase when there is no auto inducer (CAI). It phosphorylates LuxU, which transfers its phosphate to LuxO. And in our case LuxO phosphorylated activates Pqrr promoter.
Once the inducer is in the media the receptor changes to a phosphase (CqsSp) mode and the flow is reversed. LuxU is unphosphorylated and it removes the phosphate from LuxO. In consequence Pqrr promoter is repressed.
Taking this process into account and given the fact the phosphorylation process can been described as an enzymatic process; we used the Mass Action Law and the Michaelis-Menten Kinetics in order to obtain the mathematical expressions that describe the signal transduction:
Equations a and b describe the behavior of the receptor acting as a kinase (CqsS) and as a phosphase (CqsSP). The kinase is produced at a constant rate alpha CS, it is involved in Reaction 1 and has a decay rate of Gamma CS. The phosphase is also involved in Reaction 1 and has its own decay rate.
To describe the phosphorylation cascade we based our expressions on the model that Liu et al. did in 2012: LuxU and LuxO are involved in the enzymatic Reactions 2-5, both are produced by a constitute promoter at a rate a_u and a_o respectively and each molecule has its own decay rate, taking this into account Equations c-f were made.
Response →
LuxO controls Pqrr promoter. Downstream of this promoter there is the repressor TetR of the tetracycline promoter (pTet), which in our design is downstream the repressor and, controls de production of the colored response and the activator of the promoter TetA.
LuxO controls the production of TetR and it can be expressed with a Hill type equation where LuxO phosphorylated is the activator. The promoter has a small constitutive production rate alpha tr and TetR has a decay rate of gamma tr; also TetR is consumed when forms a dimer in order to repress pTet.
Equations i and j describe the production of the response and the pTet activator. As in all the preview equation, both proteins have a constitutive production and a decay rate. In our design there is a promoter (pTet) with a unique operator sequence (TetO) where the repressor (TetR2) and the activator (tTA) bind (TetR2 and tTA have the same binding domain). For that reason it was necessary to propose a mathematical expression in which the repressor and the activator compete for the binding site.
Before we established the equations above, we tried different versions of the systems. In order to get the desired response for our S. coli we needed to find the right circuit. In this section we will be showing how we did it, step by step.
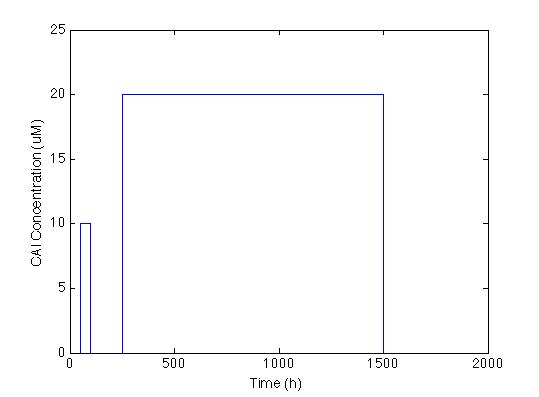
The first we did, was to establish the desired response. We want the system to react with a low and long lasting concentration of CAI. The Figure below shows the impulse we used to test the system: two pulses with different concentrations and different durations.
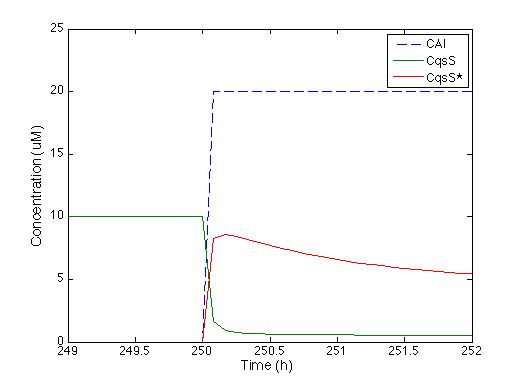
We knew that the equations of the CqsS protein behaviour are right because they were previously reported by Lelu et al (2012). So we needed to check that behaviour, the figure below shows the answer of CqsS with CAI.
As soon as the CAI molecule is present the CqsS concentration is instantly decreased, and CqsS* (which stands for the CAI bound with the Cqss protein) increases. After half an hour the concentration of CqsS starts to decrease because it is consume by LuxU,just the way it’s supposed to.
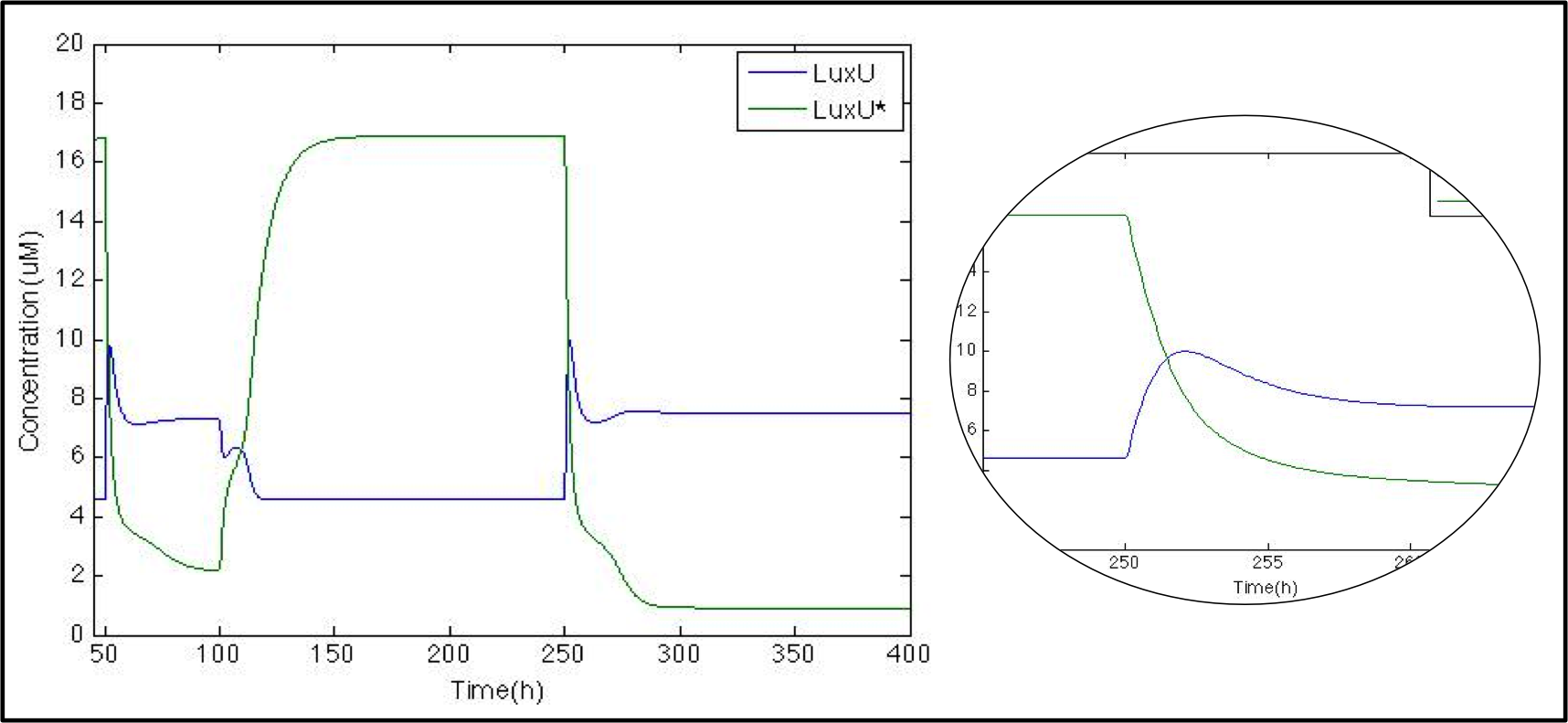
The next step is to check what happens with LuxU and its phosphorylated version now that we know that Cqss is modeled properly we want to check the phosphorylation/ phosphate cascade. As with CqsS this molecule was already modelled by Lelu et al (2012).
Just as expected whenever when CAI is present (50h-100h and 250h) LuxU is greater than LuxU* , due to the phosphase function of Cqss*. At the beginning we can see peak in LuxU concentration because of the Le Chatelier principle the reaction tends to go forward; after 3 hours the LuxU stabilises itself thanks to the phosphorylation reaction.
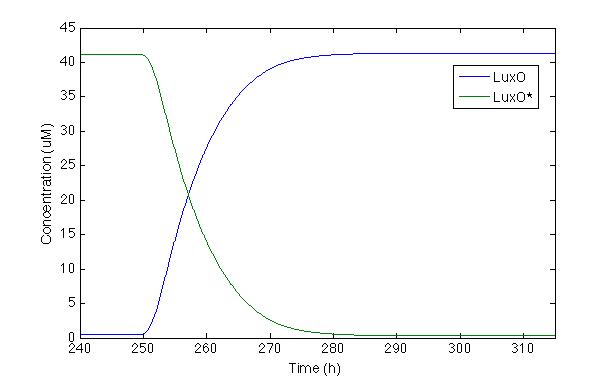
Now let’s check LuxO and its phosphorylated version. Even tough this part of the cascade was modelled by Lelu et al, we did no take into account some of the regulation routes they did, like de miRNA, because our system does not have them. We wanted to check the respond of the most important molecule from the cascade
The first time we modelled the system, LuxO response was a slower than we want. So we changed the hill constant coefficient, proposed by Lelu et al in order to make the response faster and last longer. This change compensates the lack of the negative feedback caused by miRNA that is present in V. cholera but not in S. coli.
With this modification we can see that the systems takes 20 hours to react and after 30h the production of LuxO is completely on and LuxO* is almost zero, which is desirable, because we do not want any LuxO phosphorylated to turn on the Pqrr4 promoter.
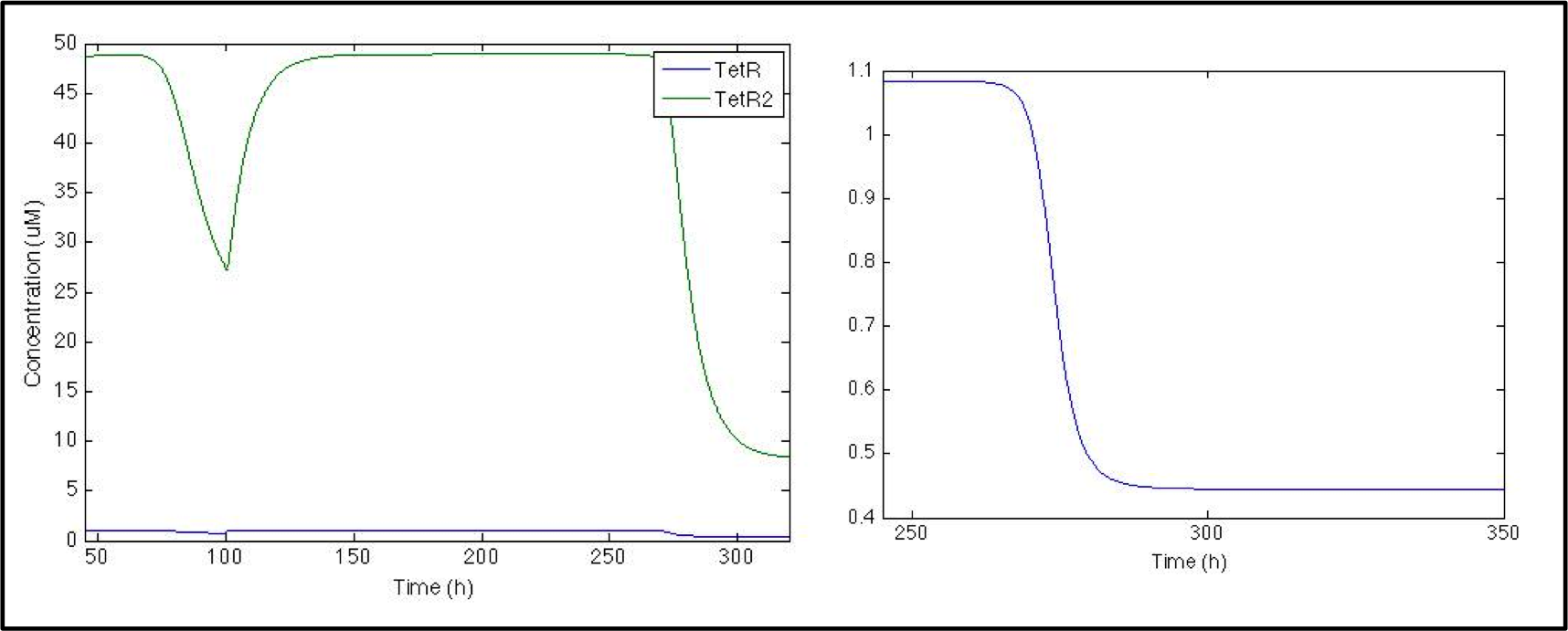
So far the model is working great, and all this part correspond to the parts registered by Peking 2011 Team. But we are really interested in the response signal and we want to see if S. coli would be able to shows us when V. cholerae is in the media. Given that when CAI is in the media de Pqrr4 promoter turns off, we need an NOT logical gate to invert the transduction, so we used the pTet and TetR system.
When CAI is present in the media TetR production turns off, which can be seen as a small change in TetR concentration; even tough the change is small it is enough to decrease the dimer concentration by 10 times. Now is time to see if this decrease is enough to turn on the production of the signal.
The first model we try was given by the equation:
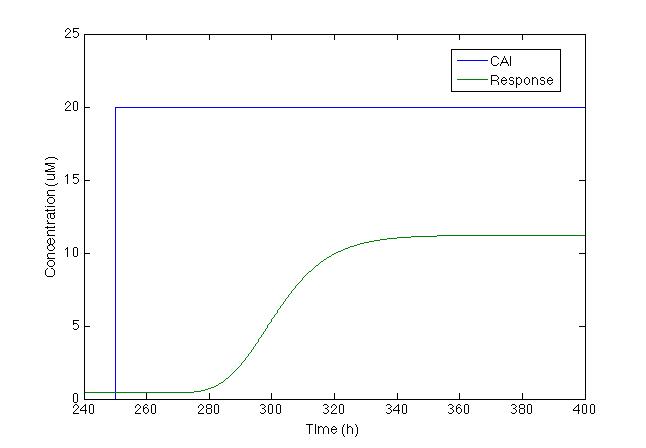
Where the production term is given by the Hill equation for repression. After running the model we got the figure below:
Even tough chromoprotein increases in 500%, the maximum concentration is less than 10uM, which makes it difficult to see without microscope. So we decided to change the model and add a positive feedback.
At first we thought of using the same promoter pTet which has an activator protein TetA . In this case the promoter and the activator will compete for the DNA binding site, so the expression for the system would be:
In order to have more options we also tried the system with another positive feedback: Psp3 with pag promoter from the Cambridge 2009 Team. In this case the activator and repressor have different DNA binding sites, so the expression for the change in the response trough time would be:
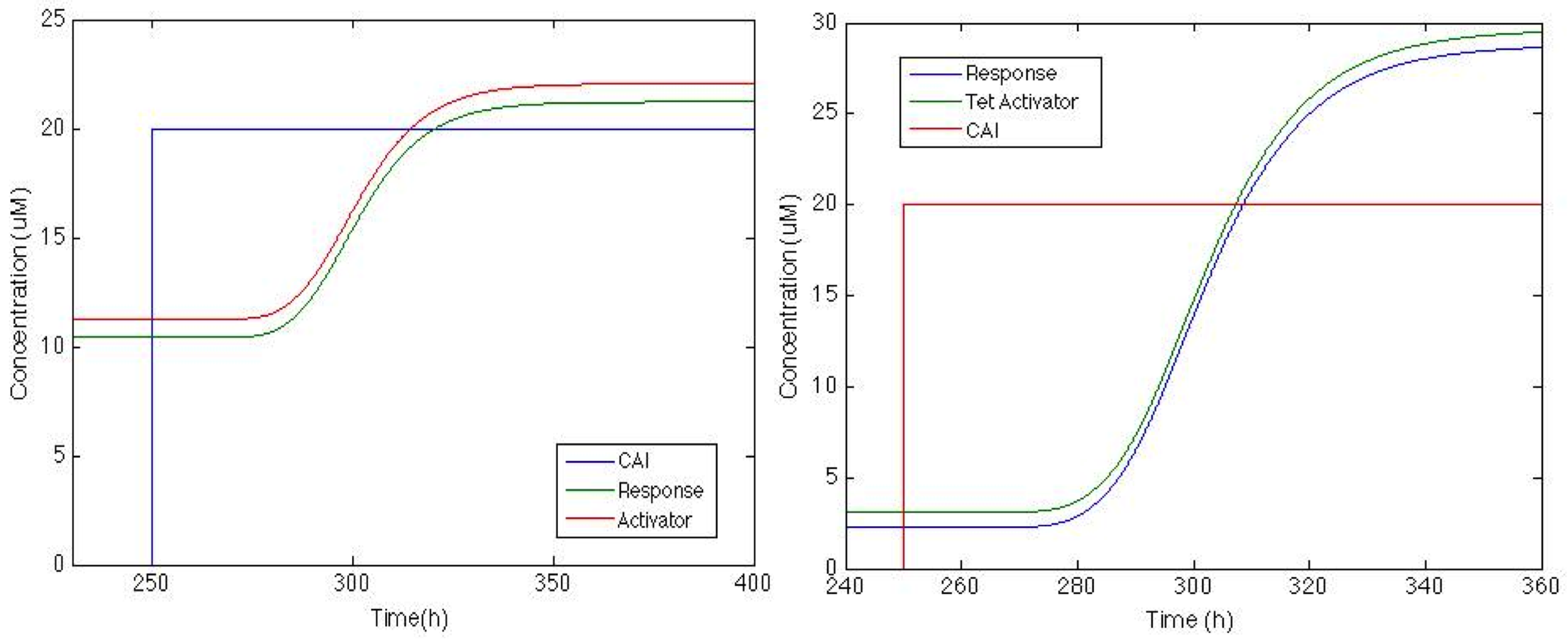
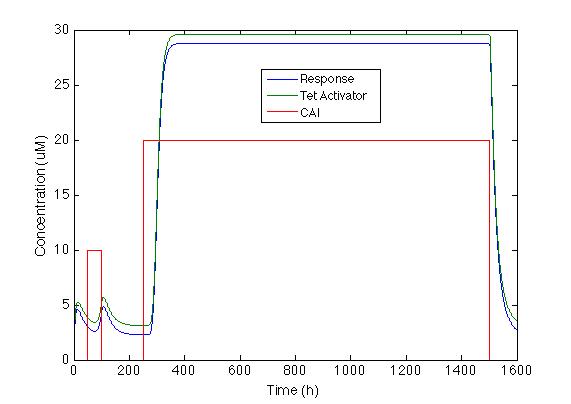
The Figure below shows how the system will behave if the positive feedback is added to the circuit. On the left is the response of Sherlock coli if the Psp3 protein is used as an activator. The chromoprotein concentration will be adequate for the system, but the increase is only about 100%.
On the other hand the response with TetA has a higher chromoprotein concentration and the increase when CAI is in the media is 12 times greater the initial concentration. This is exactly what we wanted!!!
We can see that the system takes about 20h to start producing the chromoprotein and 72h to reach its maximum expression.
 "
"