Team:Clemson
From 2014.igem.org
(→Welcome to the Official Page of Clemson iGEM!) |
|||
| (44 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{ | + | {{Template:Clemson}} |
| - | + | <html> | |
| - | < | + | <!--main content --> |
| + | <table width="70%" align="center"> | ||
| + | </html> | ||
| - | + | [[File:bumpdd.jpg|500px|frameless|center]] | |
| - | + | <html> | |
| + | <tr> | ||
| - | |||
| - | + | ||
| + | |||
| + | |||
| + | |||
| + | </td> | ||
| + | |||
| + | <tr> <td colspan="3" height="15px"> </td></tr> | ||
| + | <tr><td bgColor="#e7e7e7" colspan="3" height="1px"> </tr> | ||
| + | <tr> <td colspan="3" height="5px"> </td></tr> | ||
| + | |||
| + | |||
| + | <!--requirements section --> | ||
| + | <tr><td colspan="3"> <h3> Quorum Sensing</h3></td></tr> | ||
| + | <tr> | ||
| + | <td width="45%" valign="top"> | ||
| + | |||
| + | </html> | ||
| + | [[File:squid.jpg|500px|frameless|center]] | ||
| + | |||
| + | <br> | ||
| + | |||
| + | [[File:glowy.jpg|500px|frameless|center]] | ||
| + | <html> | ||
| + | |||
| + | </ul> | ||
| + | |||
| + | </td> | ||
| + | |||
| + | <td > </td> | ||
| + | <td width="45%"> | ||
| + | |||
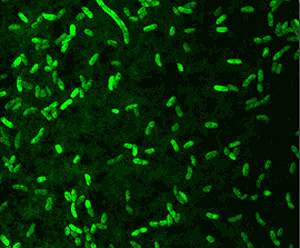
| + | <p>Quorum sensing is a means for bacteria to communicate with one another. This signaling is density-dependent, and when a large enough number of organisms are present, they even coordinate gene expression. A famous example is Vibrio fischeri, which gives the Hawaiian bobtail squid its eerie glow. When a large enough concentration of Vibrio fischeri come together in the squid's light-producing organ, or photophore, they communicate by quorum sensing and all express the luciferase gene, resulting in beautiful bioluminescence. | ||
| + | |||
| + | Quorum sensing is largely dependent upon the production of N-acyl Homoserine Lactones or AHLs. The 2014 Clemson iGEM Team worked to develop a system to detect the presence of AHLs for better study of quorum sensing and biofilm interactions. This system can be readily adapted to detect any number molecules and thus provide a flexible tool for microbiologists. </p> | ||
| + | |||
| + | </td> | ||
| + | </tr> | ||
| + | |||
| + | |||
| + | <tr> <td colspan="3" height="15px"> </td></tr> | ||
| + | <tr><td bgColor="#e7e7e7" colspan="3" height="1"> </tr> | ||
Latest revision as of 03:08, 18 October 2014
Quorum Sensing | ||
|
|
Quorum sensing is a means for bacteria to communicate with one another. This signaling is density-dependent, and when a large enough number of organisms are present, they even coordinate gene expression. A famous example is Vibrio fischeri, which gives the Hawaiian bobtail squid its eerie glow. When a large enough concentration of Vibrio fischeri come together in the squid's light-producing organ, or photophore, they communicate by quorum sensing and all express the luciferase gene, resulting in beautiful bioluminescence. Quorum sensing is largely dependent upon the production of N-acyl Homoserine Lactones or AHLs. The 2014 Clemson iGEM Team worked to develop a system to detect the presence of AHLs for better study of quorum sensing and biofilm interactions. This system can be readily adapted to detect any number molecules and thus provide a flexible tool for microbiologists. |
|
 "
"