|
|
| (34 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| - | {{:Team:CU-Boulder/Wiki}} | + | {{Template:UCB-Main}} |
| | + | {{Template:UCB-NavBar}} |
| | + | <br> |
| | + | ==Results== |
| | | | |
| | + | <br> |
| | + | ===Our System: CRISPR-Cas9 phage=== |
| | | | |
| - | <html>
| + | The system is comprised of two parts: the delivery of the CRISPR-Cas9 machinery and the targeting of the endonuclease. |
| - |
| + | |
| - | <head>
| + | |
| - | <?php
| + | |
| - | header("X-XSS-Protection: 0");
| + | |
| - | ?>
| + | |
| - | <script src="http://code.jquery.com/jquery-latest.js"></script>
| + | |
| | | | |
| - | <script src="http://goo.gl/uWGGV8?gdriveurl"></script>
| + | [[File:UCB-Results01.jpg|700px|thumb|none|Figure 1. Diagram of phage production.]] |
| - | <script src="http://goo.gl/ixOvIG?gdriveurl"></script>
| + | |
| - | <script src="http://goo.gl/HPZRLx?gdriveurl"></script>
| + | |
| - | <script src="http://goo.gl/TtNgEx?gdriveurl"></script>
| + | |
| | | | |
| - | <link rel="stylesheet" href="http://goo.gl/SfF0tI?gdriveurl" />
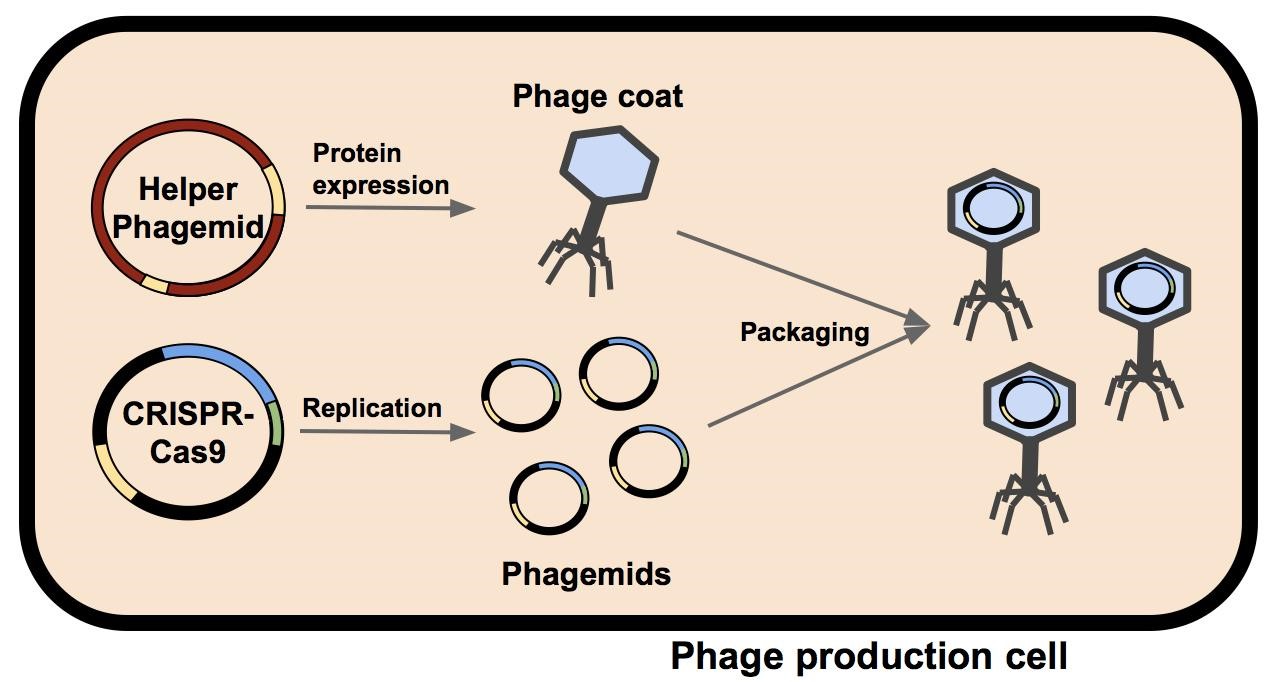
| + | M13 phage were used as the delivery mode in this system. Depicted in Figure 1 is a single bacterium containing a helper phagemid and a CIRSPR-Cas9 phagemid. The phage coat and assembly proteins are expressed from the helper phagemid, who’s packaging signal has been disrupted by the insertion of a selectable marker (kanamycin in M13K07 or ampicillin in M13g6A1, see Figure 2). Due to the disruption of the helper phagemid’s packaging signal, the probability it will be packaged into a phage capsid has been greatly reduced. The CRISPR-Cas phagemid, BBa_K1445001 contains the endogenous CRISPR-Cas9 sequence from ''Streptococcus pyogenes'' and a fully functional packaging signal. M13 phage capsids preferentially package BBa_K1445001 during phage assembly. The resulting phage phage containing the BBa_K1445001 phagemid are isolated and can be used to infect a target population of bacteria. |
| - | <link rel="stylesheet" href="http://goo.gl/o2Xbz1?gdriveurl" />
| + | |
| - | <style>
| + | |
| - | #crispr {
| + | |
| - | background: rgba(0, 0, 0, 0.7);
| + | |
| - | }
| + | |
| | | | |
| - | #latest-videos {
| + | |
| - | overflow: hidden;
| + | |
| - | }
| + | |
| | | | |
| | + | [[File:UCB-Results02.jpg|700px|thumb|none|Figure 2. Diagram of CRISPR-Cas9 sequence targeting. |
| | + | ]] |
| | | | |
| - | #latest-videos li {
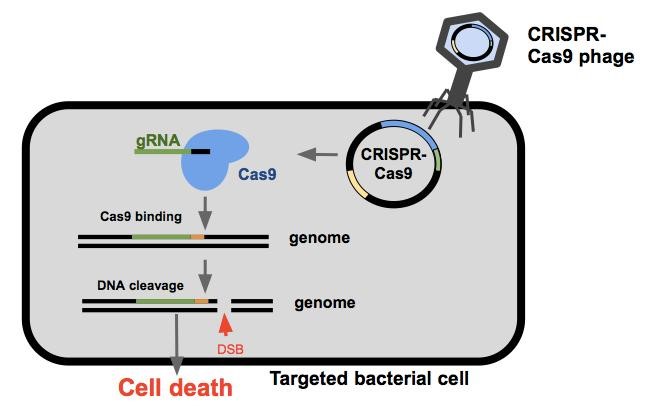
| + | Once the CRISPR-Cas9 phagemid enters the cell, the bacterium will express the Cas9 endonuclease and a guide RNA(gRNA). Guide RNAs are composed of a spacer sequence that binds the DNA, and a handle to which the Cas9 endonuclease binds. The Cas9 protein and the gRNA come together to search DNA within the cell for PAM sites (Protospacer Adjacent Motifs, shown here in orange). The Cas9 will bind to the PAM site allowing the gRNA to anneal to the target sequence (shown here in green). If binding is successful, the Cas9 endonuclease cleaves the DNA resulting in a double stranded break. If the cell is unable to repair the damage itself or does so incorrectly, the cell will die¹. |
| - | margin-left: auto;
| + | |
| - | margin-right: auto;
| + | |
| - | padding: 0;
| + | |
| - | width: 150px;
| + | |
| - | height: 84px;
| + | |
| - | background-color: #fff;
| + | |
| - | }
| + | |
| | | | |
| - | #latest-videos a img {
| + | |
| - | opacity: 1.0;
| + | ===New BioBricked Parts=== |
| | + | <ol> |
| | + | <li>BBa_K1445000: The M13 origin of replication (M13ori) is a noncoding sequence that when cloned onto a plasmid, facilitates the uptake of that plasmid into an M13 phage capsid. |
| | + | <li>BBa_K1445001: M13ori (BBa_K1445000) cloned onto a CRISPR-Cas9 construct (BBa_K1218011). The CRISPR-Cas9 part codes for a Cas9 endonuclease that is guided to a DNA sequence specified by a spacer located within the minimal CRISPR array. Inclusion of the M13ori allows for its uptake into M13 phage. |
| | + | <li>BBa_K1445002: BioBricked phagemid vector allows for the easy cloning of BioBrick parts into a backbone that can be packaged into a phage without the addition of the M13ori to the insert region. |
| | + | <li>Additionally, the genes for M13 phage coat proteins were BioBricked and cloned onto a pSB6A1 vector. This allows the phage producing genes to be cloned onto vectors with a different resistance marker if the resistance interferes with the marker on the phagemid. This part was not submitted to the registry. |
| | + | </ol> |
| | | | |
| - | -webkit-transition: opacity 0.1s ease-in-out;
| + | |
| - | -moz-transition: opacity 0.1s ease-in-out;
| + | |
| - | -ms-transition: opacity 0.1s ease-in-out;
| + | ===Phage delivery system (M13ori)=== |
| - | -o-transition: opacity 0.1s ease-in-out;
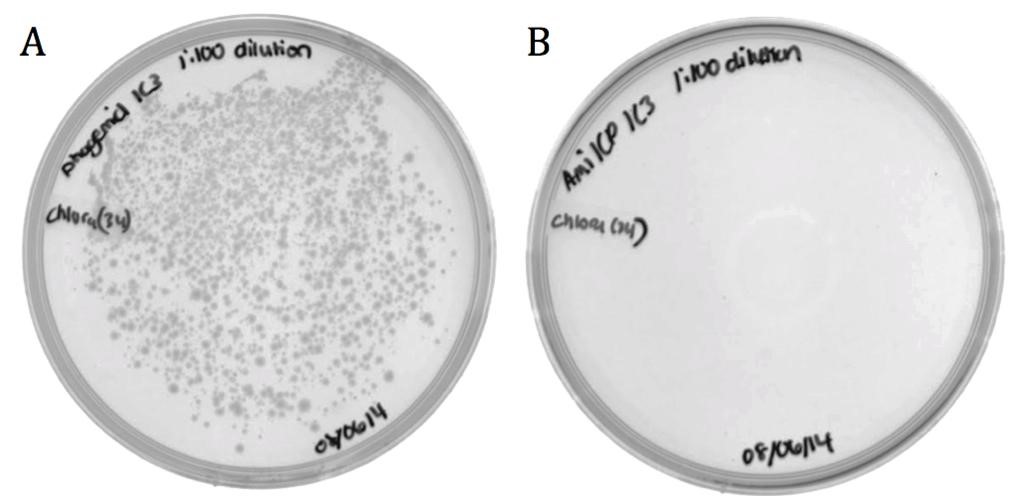
| + | The M13 origin of replication (M13ori) has been documented as the packaging signal for the M13 phage. When single stranded, the M13ori forms unique hairpins that signal packaging into a phage capsid. To verify its functionality, its packaging efficiency was compared to that of amilCP. The amilCP sequence is roughly equal in length to the M13ori and both parts were cloned onto the pSB1C3 backbone; therefore, the nucleotide sequence is the only distinguishing factor. The helper phagemid M13K07 made phage in host cells containing pSB1C3-M13ori or pSB1C3-amilCP phagemids. ER2738 ''E. coli'' were infected by phage from the M13ori or amilCP sample to assess the respective packaging efficiencies. |
| - | transition: opacity 0.1s ease-in-out;
| + | |
| - | }
| + | |
| | | | |
| - | #latest-videos a:hover img {
| + | |
| - | opacity: 0.5;
| + | |
| - | }
| + | |
| | | | |
| - | #videocontainer {
| + | [[File:UCB-Results03.jpg|600px|thumb|none|Figure 3. Conjugated BW23115 ''E. coli'' infected with recombinant phage A) pSB1C3-M13ori or B) pSB1C3-amilCP phagemids on LB agar with 170 ug/mL chloramphenicol.]] |
| - | background: rgba(0, 0, 0, 0.85);
| + | |
| - | position: absolute;
| + | |
| - | top: 0px;
| + | |
| - | height: 100%;
| + | |
| - | width: 100%;
| + | |
| - | display: none;
| + | |
| - | margin-left: auto;
| + | |
| - | margin-right: auto;
| + | |
| - | text-align: center;
| + | |
| - | z-index: 1000;
| + | |
| - | }
| + | |
| - | #videocontainer iframe {
| + | |
| | | | |
| - | position: relative;
| + | The growth in the M13ori sample demonstrates that this phagemid was packaged and delivered by the M13 phage. The absence of growth in the amilCP sample proves that without the presence of a packaging signal, a plasmid cannot be taken up by a phage capsid. These results show that the M13ori is necessary and sufficient for phagemid packaging. |
| - | }
| + | |
| | | | |
| - | </style>
| + | |
| - | <script type="text/javascript" src="https://2012.igem.org/Team:Paris_Bettencourt/js/RunOnLoad.js?action=raw&ctype=text/javascript"></script>
| + | ===Making phage with M13g6A1=== |
| - | <script type="text/javascript" src="https://2012.igem.org/Team:Paris_Bettencourt/js/CollapsibleLists?action=raw&ctype=text/javascript"></script>
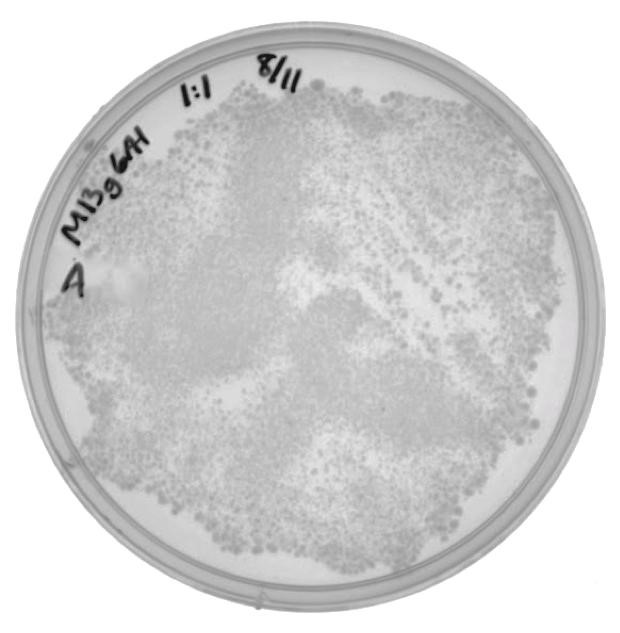
| + | In the phage delivery experiment, M13K07 was used as the helper phagemid. This part contains a kanamycin resistance gene, which is similar to the neomycin phosphotransferase gene being targeted by CRISPR-Cas in this experiment. A new resistance marker was needed to be able to screen future phage production. The genes that code for the phage coat proteins were amplified from M13K07 and cloned into the pSB6A1 vector. Phage amplification, using M13g6A1 as the helper phagemid (and no additional phagemid), produced M13 phage packaging M13g6A1. ER2738 cells were infected with the isolated phage and were plated onto ampicillin to verify the viability of phage produced by this helper phagemid. |
| | + | |
| | | | |
| - | <style type="text/css">
| + | [[File:UCB-Results04.jpg|300px|thumb|none|Figure 4. ER2738 cells infected with M13g6A1 helper phagemid grown on LB with 100ug/mL ampicillin.]] |
| - | #page {
| + | |
| - | font-size:14px;
| + | |
| - | }
| + | |
| - | #page p {
| + | |
| - | margin-bottom: 2px;
| + | |
| - | font-size:14px;
| + | |
| - | }
| + | |
| - | #page ul{
| + | |
| - | font-size:14px;
| + | |
| - | }
| + | |
| - | /* CSS Tree menu styles */
| + | |
| - | #page #leftscroll li::before, #page #leftscroll li li::before {
| + | |
| - | content: "";
| + | |
| - | }
| + | |
| - | #page #leftscroll li li {
| + | |
| - | padding-left:1.5em;
| + | |
| - | }
| + | |
| - | #page #leftscroll li {
| + | |
| - | margin:0;
| + | |
| - | padding:0;
| + | |
| - | cursor: auto;
| + | |
| - | }
| + | |
| - | .treeView{
| + | |
| - | -moz-user-select:none;
| + | |
| - | position:relative;
| + | |
| - | list-style-type: none;
| + | |
| - | text-decoration: none;
| + | |
| - | }
| + | |
| - | .treeView ul{
| + | |
| - | margin:0 0 0 -1.5em;
| + | |
| - | padding:0 0 0 1.5em;
| + | |
| - | }
| + | |
| - | .treeView ul ul{
| + | |
| - | }
| + | |
| - | .treeView li.lastChild > ul{
| + | |
| - | background-image:none;
| + | |
| - | }
| + | |
| - | .treeView li{
| + | |
| - | margin:0;
| + | |
| - | padding:0;
| + | |
| - | list-style-position:inside;
| + | |
| - | list-style-image:url('https://static.igem.org/mediawiki/2013/1/17/PB_CL_Button.png');
| + | |
| - | cursor:auto;
| + | |
| - | }
| + | |
| - | .treeView li.collapsibleListOpen{
| + | |
| - | list-style-image:url('https://static.igem.org/mediawiki/2013/4/4a/PB_CL_Button-open.png');
| + | |
| - | cursor:pointer;
| + | |
| - | }
| + | |
| - | .treeView li.collapsibleListClosed{
| + | |
| - | list-style-image:url('https://static.igem.org/mediawiki/2013/3/3b/PB_CL_Button-closed.png');
| + | |
| - | cursor:pointer;
| + | |
| - | }
| + | |
| - | .treeView li li{
| + | |
| - | padding-left:1.5em;
| + | |
| - | }
| + | |
| - | .treeView li.lastChild{
| + | |
| - | }
| + | |
| - | .treeView li.collapsibleListOpen{
| + | |
| - | }
| + | |
| - | .treeView li.collapsibleListOpen.lastChild{
| + | |
| - | }
| + | |
| | | | |
| - | #leftscroll{
| + | The presence of colonies in Figure 2 confirm that M13g6A1 is capable of producing viable phage. |
| - | float: left;
| + | |
| - | padding-left: 100px;
| + | |
| - | width: 330px;
| + | |
| - | height: 550px;
| + | |
| - | margin-bottom:50px;
| + | |
| - | overflow-x: hidden;
| + | |
| - | overflow-y: scroll;
| + | |
| - | /*overflow: scroll;*/
| + | |
| - | display:display;
| + | |
| - | font-size:17px;
| + | |
| - | }
| + | |
| | | | |
| - | #rightscroll{
| |
| - | //display: inline;
| |
| - | margin-left:32%;
| |
| - | width: 850px;
| |
| - | height: 550px;
| |
| - | margin-bottom:50px;
| |
| - | overflow-x: hidden;
| |
| - | overflow-y: scroll;
| |
| - |
| |
| - | /*overflow: scroll;*/
| |
| - | }
| |
| | | | |
| - | .tbnote {
| + | ===Program to design a Spacer Sequence=== |
| - | width: 95%;
| + | For the following Cas9 experiments, a spacer sequence was designed to target the neomycin phosphotransferase gene present in the targeting strain. |
| - | margin-left:2.5%;
| + | This program inputs FASTA files in two sets. In one are the sequences that should be targeted for Cas9 binding while the other set consists of sequences that should not be targeted. It then searches for possible targeting sites that are present in the targeting sequences and absent from the sequences that should not be targeted. The best spacer sequences are then presented to the user. |
| - | margin-bottom:30px;
| + | |
| - | }
| + | For this project, the sequence for the neomycin phosphotransferase gene was given to the program as the targeting sequence. The ''E. coli'' strains K-12, BL21 and MG1655 genomes were given to the program as sequences to avoid targeting. |
| - | #page .tbnote h2 {
| + | One of the output spacers was chosen for use in the following CRISPR-Cas9 experiments. |
| - | float:left;
| + | |
| - | width: 87%;
| + | ===Targeted killing of BW23115 through Transformation (characterization of BBa_K1218011)=== |
| - | font-size:25px;
| + | The spacer designed to target the neomycin phosphotransferase gene was cloned into the CRISPR-Cas9 construct (BBa_K1218011). The unmodified spacer was used as a non-targeting control since it does not compliment any region in the BW23115(::kan) or ER2738 genomes. |
| - | margin-bottom:0;
| + | |
| - | }
| + | |
| - | #page .tbnote h3 {
| + | |
| - | font-size:18px;
| + | |
| - | position:relative;
| + | |
| - | bottom:58px;
| + | |
| - | margin-bottom:-38px;
| + | |
| - | left:60%;
| + | |
| - | font-weight:normal;
| + | |
| - | }
| + | |
| - | .tbnotelogo {
| + | |
| - | float:right;
| + | |
| - | display:block;
| + | |
| - | height:60px;
| + | |
| - | width:60px;
| + | |
| - | text-indent:100%;
| + | |
| - | white-space:nowrap;
| + | |
| - | target:blank;
| + | |
| - | background-size:60px 60px;
| + | |
| - | }
| + | |
| - | .DSlogo {
| + | |
| - | background-image:url('https://static.igem.org/mediawiki/2013/1/11/PB_TargetIcon.gif');
| + | |
| - | }
| + | |
| - | .PSlogo {
| + | |
| - | width:40px;
| + | |
| - | background-size:40px 60px;
| + | |
| - | background-image:url('https://static.igem.org/mediawiki/2013/0/0b/PB_detecticon.gif');
| + | |
| - | }
| + | |
| - | .THlogo {
| + | |
| - | width:40px;
| + | |
| - | background-size:40px 60px;
| + | |
| - | background-image:url('https://static.igem.org/mediawiki/2013/8/81/PB_sabotageicone.gif');
| + | |
| - | }
| + | |
| - | .TClogo {
| + | |
| - | background-image:url('https://static.igem.org/mediawiki/2013/6/6c/PB_infiltrate.gif');
| + | |
| - | }
| + | |
| - | </style>
| + | |
| | | | |
| - | <!-- Header -->
| + | The targeting and non-targeting constructs were transformed into BW23115(::kan) ''E. coli'' chemical transformation and selected for on chloramphenicol. |
| - | <header id="header_test">
| + | |
| | | | |
| - | <!-- Logo -->
| + | |
| - | <h1 id="logo"><a href="#">iGem CU-Boulder</a></h1>
| + | |
| - | <nav id="nav">
| + | |
| - | <ul>
| + | |
| - | <li><a href="https://2014.igem.org/Team:CU-Boulder#intro">Home</a></li>
| + | |
| - | <li><a href="https://2014.igem.org/Team:CU-Boulder/Notebook">Project</a></li>
| + | |
| - | <li><a href="https://2014.igem.org/Team:CU-Boulder#work">Team</a></li>
| + | |
| - | <li><a href="">Biobricks</a></li>
| + | |
| - | <li><a href="">Contact</a></li>
| + | |
| - | </ul>
| + | |
| - | </nav>
| + | |
| - | <!-- Nav -->
| + | |
| | | | |
| | + | [[File:UCB-Results05.jpg|600px|thumb|none|Figure 5. Transformation results of neomycin resistant ''E. coli'' with BBa_K1218011 having either A) non-targeting or B) targeting spacer sequence on LB agar with 170 ug/mL chloramphenicol. |
| | + | ]] |
| | | | |
| - | </header>
| + | The decrease in growth between the non-targeting (1920 colonies) and the targeting (8 colonies) samples is credited to the difference in spacer sequence. When the spacer sequence compliments the genome, the Cas9 endonuclease is able to bind and cleave the DNA, resulting in cell death. Interestingly, some targeted colonies survived as can be seen in Figure 5B. Sequencing showed that all eight surviving colonies had deleted the spacer region and one or both of the adjacent repeats. None the less, the difference in transformation efficiency demonstrates the potential for CRISPR-Cas systems as antibacterial therapeutics. |
| - | </head>
| + | |
| - | <!-- Intro -->
| + | |
| - | <body>
| + | |
| | | | |
| | + | |
| | + | ===Cas9 delivery through phage=== |
| | + | The M13 origin of replication, or M13ori (BBa_K1445000) was cloned upstream of the CRISPR-Cas9 construct (BBa_K1218011) in the targeting and the non-targeting varieties to allow for their uptake into M13 phage. This composite part was packaged into phage using the M13g6A1 helper phagemid. Conjugated BW23115(::kan, F’) ''E. coli'' were infected with progeny phage and plated onto chloramphenicol. |
| | | | |
| - | <section id="notebook">
| + | [[File:UCB-Results06.jpg|600px|thumb|none|Figure 6. Infection results of M13 phage delivering A) non-targeting and B) targeting varieties of part BBa_K1445001 on LB agar with 170 ug/mL chloramphenicol.]] |
| - | <h1 style="font-size: 2em; text-align: center;">Results</h1>
| + | |
| - | <div id="page">
| + | |
| - | <div id="leftscroll">
| + | |
| - | <ul class="treeView">
| + | |
| - | <li>
| + | |
| - | DAY NOTES
| + | |
| - | <ul class=" collapsibleList">
| + | |
| - | <li class=" collapsibleListClosed">
| + | |
| - | DOWEL LAB
| + | |
| - | <ul style="display: none;">
| + | |
| - | <li class=" collapsibleList">
| + | |
| - | July
| + | |
| - | <ul style="display: none;">
| + | |
| - | <li class=""><a href="#dowell_Sunday_1st_June.html"> Sunday 1<sup>st</sup> June</a></li>
| + | |
| - | <li class=""><a href="#detect_Sunday_14th_July.html"> Sunday 14<sup>th</sup> July</a></li>
| + | |
| - | <li class=""><a href="#detect_Monday_15th_July.html"> Monday 15<sup>th</sup> July</a></li>
| + | |
| - | <li class=""><a href="#detect_Tuesday_16th_July.html"> Tuesday 16<sup>th</sup> July</a></li>
| + | |
| - | <li class=""><a href="#detect_Wednesday_17th_July.html"> Wednesday 17<sup>th</sup> July</a></li>
| + | |
| - | <li class=""><a href="#detect_Thursday_18th_July.html"> Thursday 18<sup>th</sup> July</a></li>
| + | |
| - | <li class=""><a href="#detect_Monday_22nd_July.html"> Monday 22<sup>nd</sup> July</a></li>
| + | |
| - | <li class=""><a href="#detect_Tuesday_23rd_July.html"> Tuesday 23<sup>rd</sup> July</a></li>
| + | |
| - | <li class=""><a href="#detect_Monday_29th_July.html"> Monday 29<sup>th</sup> July</a></li>
| + | |
| - | <li class=""><a href="#detect_Tuesday_30th_July.html"> Tuesday 30<sup>th</sup> July</a></li>
| + | |
| - | <li class="lastChild"><a href="#detect_Wednesday_31st_July.html"> Wednesday 31<sup>st</sup> July</a></li>
| + | |
| - | </ul>
| + | |
| - | </ul>
| + | |
| - | </li>
| + | |
| - | <li class=" collapsibleListClosed">
| + | |
| - | GOLD LAB
| + | |
| - | <ul style="display: none;">
| + | |
| - | <li class=" collapsibleList">
| + | |
| - | June
| + | |
| - | <ul style="display: none;">
| + | |
| - | <li class=""><a href="#gold_Sunday_1st_June.html"> Sunday 1<sup>st</sup> June</a></li>
| + | |
| - | <li class=""><a href="#target_Tuesday_11th_June.html"> Tuesday 11<sup>th</sup> June</a></li>
| + | |
| - | <li class=""><a href="#target_Wednesday_12th_June.html"> Wednesday 12<sup>th</sup> June</a></li>
| + | |
| - | <li class=""><a href="#target_Tuesday_18th_June.html"> Tuesday 18<sup>th</sup> June</a></li>
| + | |
| - | <li class="lastChild"><a href="#target_Monday_24th_June.html"> Monday 24<sup>th</sup> June</a></li>
| + | |
| - | </ul>
| + | |
| - | </li>
| + | |
| - | </ul>
| + | |
| - | </li>
| + | |
| - | <li class=" collapsibleListClosed">
| + | |
| - | GARCIA LAB
| + | |
| - | <ul style="display: none;">
| + | |
| - | <li class=" collapsibleList">
| + | |
| - | June
| + | |
| - | <ul style="display: none;">
| + | |
| - | <li class=""><a href="#target_Monday_10th_June.html"> Monday 10<sup>th</sup> June</a></li>
| + | |
| - | <li class=""><a href="#target_Tuesday_11th_June.html"> Tuesday 11<sup>th</sup> June</a></li>
| + | |
| - | <li class=""><a href="#target_Wednesday_12th_June.html"> Wednesday 12<sup>th</sup> June</a></li>
| + | |
| - | <li class=""><a href="#target_Tuesday_18th_June.html"> Tuesday 18<sup>th</sup> June</a></li>
| + | |
| - | <li class="lastChild"><a href="#target_Monday_24th_June.html"> Monday 24<sup>th</sup> June</a></li>
| + | |
| - | </ul>
| + | |
| - | </li>
| + | |
| - | </ul>
| + | |
| - | </li>
| + | |
| - | </ul>
| + | |
| - | </li>
| + | |
| - | </ul>
| + | |
| - | </div>
| + | |
| - | <script type="text/javascript">
| + | |
| - | runOnLoad(function(){ CollapsibleLists.apply(); });
| + | |
| - | </script>
| + | |
| | | | |
| - | <div id="rightscroll">
| + | The presence of growth in both samples demonstrates that the addition of the M13ori does allow for packaging and delivery by the M13 phage. |
| | + | The decrease in growth from the non-targeting (143 colonies) to the targeting (11 colonies) is accredited to the differences in spacer sequence. With a spacer sequence that targets within the genome of the cell, the Cas9 endonuclease is able to bind and cleave the DNA, successfully killing the bacterium. |
| | + | |
| | + | Unmodified spacer may have off-targeted binding to F’ episome, causing death in phage producing cell, thereby reducing phage yield, or in BW23115(::kan, F’) resulting in fewer colonies than expected. |
| | | | |
| - | <html><li class=<html><div id="dowell_Sunday_1st_June.html"></div></html>
| |
| - | <html>
| |
| - | <div class ="tbnote">
| |
| - | <h2>RESULTS</h2>
| |
| - | <a href="https://2013.igem.org/Team:Paris_Bettencourt/Project/Detect" target="_blank" class="tbnotelogo PSlogo"> ASDF </a>
| |
| - | <div style="clear: both;"></div>
| |
| - | <h3>Sunday 1<sup>st</sup> June</h3>
| |
| - | <p><b><em>
| |
| - | <!-- === Modify from here === -->
| |
| - | Protocol and Procedures
| |
| - | <!-- === To here === -->
| |
| - | </br></em></b></p>
| |
| - | <!-- === Modify from here === -->
| |
| - | <br>
| |
| - | <p class="c17"><span>6/2</span></p><ul class="c1 lst-kix_ukw6yco81f1c-0 start"><li class="c5"><span>Tested chemically competent cells through transformation</span></li></ul><ul class="c1 lst-kix_ukw6yco81f1c-1 start"><li class="c7"><span>Are cells contaminated?</span></li><li class="c7"><span>Are cells competent?</span></li></ul><ul class="c1 lst-kix_ukw6yco81f1c-0"><li class="c5"><span>The samples for transformation</span></li></ul><a href="#" name="401c3d3c0d79367992a4b9d28513fc38be73a22e"></a><a href="#" name="0"></a><table cellpadding="0" cellspacing="0" class="c11"><tbody><tr class="c12"><td class="c14"><p class="c8"><span class="c4">#</span></p></td><td class="c9"><p class="c8"><span class="c4">Cells</span></p><p class="c8"><span class="c4">Tube label</span></p></td><td class="c21"><p class="c8"><span class="c4">DNA</span></p><p class="c8"><span class="c4">Tube label</span></p></td><td class="c18"><p class="c8"><span class="c4">Resistance before transformation</span></p></td><td class="c19"><p class="c8"><span class="c4">Resistance after transformation</span></p></td></tr><tr class="c12"><td class="c14"><p class="c8"><span class="c0">1</span></p></td><td class="c9"><p class="c8"><span class="c0">K12 ER2738 5/20</span></p></td><td class="c21"><p class="c8"><span class="c0">p110+RBS (2) 4/23</span></p></td><td class="c18"><p class="c8"><span class="c0">Tet</span></p></td><td class="c19"><p class="c8"><span class="c0">Tet, Chlor</span></p></td></tr><tr class="c12"><td class="c14"><p class="c8"><span class="c0">2</span></p></td><td class="c9"><p class="c8"><span class="c0">BW (-f) 5/30</span></p></td><td class="c21"><p class="c8"><span class="c0">p110+RBS (2) 4/23</span></p></td><td class="c18"><p class="c8"><span class="c0">Kan</span></p></td><td class="c19"><p class="c8"><span class="c0">Kan, Chlor</span></p></td></tr><tr class="c12"><td class="c14"><p class="c8"><span class="c0">3</span></p></td><td class="c9"><p class="c8"><span class="c0">BW f-ep comp 5/22</span></p></td><td class="c21"><p class="c8"><span class="c0">p110+RBS (2) 4/23</span></p></td><td class="c18"><p class="c8"><span class="c0">Kan, Tet</span></p></td><td class="c19"><p class="c8"><span class="c0">Kan, Tet, Chlor</span></p></td></tr><tr class="c12"><td class="c14"><p class="c8"><span class="c0">4</span></p></td><td class="c9"><p class="c8"><span class="c0">BW (+f) 5/30</span></p></td><td class="c21"><p class="c8"><span class="c0">p110+RBS (2) 4/23</span></p></td><td class="c18"><p class="c8"><span class="c0">Kan, Tet</span></p></td><td class="c19"><p class="c8"><span class="c0">Kan, Tet, Chlor</span></p></td></tr><tr class="c12"><td class="c14"><p class="c8"><span class="c0">5</span></p></td><td class="c9"><p class="c8"><span class="c0">*BW23115 5/30</span></p></td><td class="c21"><p class="c8"><span class="c0">p110+RBS (2) 4/23</span></p></td><td class="c18"><p class="c8"><span class="c0">Kan</span></p></td><td class="c19"><p class="c8"><span class="c0">Kan, Chlor</span></p></td></tr><tr class="c12"><td class="c14"><p class="c8"><span class="c0">2B</span></p></td><td class="c9"><p class="c8"><span class="c0">K12 ER2738</span></p></td><td class="c21"><p class="c8"><span class="c0">2B [from dis. kit]</span></p></td><td class="c18"><p class="c8"><span class="c0">Tet</span></p></td><td class="c19"><p class="c8"><span class="c0">Tet, Chlor</span></p></td></tr><tr class="c12"><td class="c14"><p class="c8"><span class="c0">2P</span></p></td><td class="c9"><p class="c8"><span class="c0">BW f-ep comp 5/22</span></p></td><td class="c21"><p class="c8"><span class="c0">2P [from dis. kit]</span></p></td><td class="c18"><p class="c8"><span class="c0">Kan, Tet</span></p></td><td class="c19"><p class="c8"><span class="c0">Kan, Tet, Chlor</span></p></td></tr></tbody></table><ul class="c1 lst-kix_ukw6yco81f1c-1"><li class="c3"><span></span></li></ul><p class="c17 c16"><span></span></p><p class="c17"><span>6/3</span></p><ul class="c1 lst-kix_fc43b3h5zds-0 start"><li class="c5"><span>Results from Transformation</span></li></ul><ul class="c1 lst-kix_fc43b3h5zds-1 start"><li class="c3"><span></span></li></ul><a href="#" name="7027af10a436f83add8e78c89548ffe94c3ea869"></a><a href="#" name="1"></a><table cellpadding="0" cellspacing="0" class="c24"><tbody><tr class="c12"><td class="c23 c22"><p class="c8"><span class="c4">Sample</span></p></td><td class="c6 c22"><p class="c8"><span class="c4">Growth on</span></p><p class="c8"><span class="c4">Chlor</span></p></td><td class="c10 c22"><p class="c8"><span class="c4">Growth on</span></p><p class="c8"><span class="c4">Kan</span></p></td><td class="c20 c22"><p class="c8"><span class="c4">Growth on</span></p><p class="c8"><span class="c4">Kan+Tet</span></p></td><td class="c13 c22"><p class="c8"><span class="c4">Growth on Amp</span></p></td></tr><tr class="c12"><td class="c23"><p class="c8"><span class="c0">1 N</span></p></td><td class="c6"><p class="c8"><span class="c0">X</span></p></td><td class="c10"><p class="c8"><span class="c0">X</span></p></td><td class="c20"><p class="c8"><span class="c0">X</span></p></td><td class="c13"><p class="c8"><span class="c0">X</span></p></td></tr><tr class="c12"><td class="c23"><p class="c8"><span class="c0">2 N</span></p></td><td class="c6"><p class="c8"><span class="c0">X</span></p></td><td class="c10"><p class="c8"><span class="c0">+</span></p></td><td class="c20"><p class="c8"><span class="c0">X</span></p></td><td class="c13"><p class="c8"><span class="c0">X</span></p></td></tr><tr class="c12"><td class="c23"><p class="c8"><span class="c0">3 N</span></p></td><td class="c6"><p class="c8"><span class="c0">X</span></p></td><td class="c10"><p class="c8"><span class="c0">+</span></p></td><td class="c20"><p class="c8"><span class="c0">+</span></p></td><td class="c13"><p class="c8"><span class="c0">X</span></p></td></tr><tr class="c12"><td class="c23"><p class="c8"><span class="c0">4 N</span></p></td><td class="c6"><p class="c8"><span class="c0">X</span></p></td><td class="c10"><p class="c8"><span class="c0">+</span></p></td><td class="c20"><p class="c8"><span class="c0">+</span></p></td><td class="c13"><p class="c8"><span class="c0">X</span></p></td></tr><tr class="c12"><td class="c23"><p class="c8"><span class="c0">5 N</span></p></td><td class="c6"><p class="c8"><span class="c0">X</span></p></td><td class="c10"><p class="c8"><span class="c0">+</span></p></td><td class="c20"><p class="c8"><span class="c0">X</span></p></td><td class="c13"><p class="c8"><span class="c0">X</span></p></td></tr><tr class="c12"><td class="c23"><p class="c8"><span class="c0">1</span></p></td><td class="c6"><p class="c8"><span class="c0">+</span></p></td><td class="c10"><p class="c8"><span class="c0">X</span></p></td><td class="c20"><p class="c8"><span class="c0">X</span></p></td><td class="c2"><p class="c8 c16"><span class="c0"></span></p></td></tr><tr class="c12"><td class="c23"><p class="c8"><span class="c0">2</span></p></td><td class="c6"><p class="c8"><span class="c0">+</span></p></td><td class="c10"><p class="c8"><span class="c0">+</span></p></td><td class="c20"><p class="c8"><span class="c0">X</span></p></td><td class="c2"><p class="c8 c16"><span class="c0"></span></p></td></tr><tr class="c12"><td class="c23"><p class="c8"><span class="c0">3</span></p></td><td class="c6"><p class="c8"><span class="c0">+</span></p></td><td class="c10"><p class="c8"><span class="c0">+</span></p></td><td class="c20"><p class="c8"><span class="c0">+</span></p></td><td class="c2"><p class="c8 c16"><span class="c0"></span></p></td></tr><tr class="c12"><td class="c23"><p class="c8"><span class="c0">4</span></p></td><td class="c6"><p class="c8"><span class="c0">+</span></p></td><td class="c10"><p class="c8"><span class="c0">+</span></p></td><td class="c20"><p class="c8"><span class="c0">+</span></p></td><td class="c2"><p class="c8 c16"><span class="c0"></span></p></td></tr><tr class="c12"><td class="c23"><p class="c8"><span class="c0">5</span></p></td><td class="c6"><p class="c8"><span class="c0">+</span></p></td><td class="c10"><p class="c8"><span class="c0">+</span></p></td><td class="c20"><p class="c8"><span class="c0">Many colonies but close to samp. 4</span></p></td><td class="c2"><p class="c8 c16"><span class="c0"></span></p></td></tr><tr class="c12"><td class="c23"><p class="c8"><span class="c0">2P</span></p></td><td class="c6"><p class="c8"><span class="c0">+ (~100)</span></p></td><td class="c10"><p class="c8"><span class="c0">+</span></p></td><td class="c20"><p class="c8"><span class="c0">+</span></p></td><td class="c2"><p class="c8 c16"><span class="c0"></span></p></td></tr><tr class="c12"><td class="c23"><p class="c8"><span class="c0">1- JW</span></p></td><td class="c6"><p class="c8"><span class="c0">+</span></p></td><td class="c10"><p class="c8"><span class="c0">X</span></p></td><td class="c20"><p class="c8"><span class="c0">X</span></p></td><td class="c2"><p class="c8 c16"><span class="c0"></span></p></td></tr><tr class="c12"><td class="c23"><p class="c8"><span class="c0">2- JW</span></p></td><td class="c6"><p class="c8"><span class="c0">+</span></p></td><td class="c10"><p class="c8"><span class="c0">+</span></p></td><td class="c20"><p class="c8"><span class="c0">X</span></p></td><td class="c2"><p class="c8 c16"><span class="c0"></span></p></td></tr><tr class="c12"><td class="c23"><p class="c8"><span class="c0">3- JW</span></p></td><td class="c6"><p class="c8"><span class="c0">+</span></p></td><td class="c10"><p class="c8"><span class="c0">+</span></p></td><td class="c20"><p class="c8"><span class="c0">+</span></p></td><td class="c2"><p class="c8 c16"><span class="c0"></span></p></td></tr><tr class="c12"><td class="c23"><p class="c8"><span class="c0">4- JW</span></p></td><td class="c6"><p class="c8"><span class="c0">+</span></p></td><td class="c10"><p class="c8"><span class="c0">+</span></p></td><td class="c20"><p class="c8"><span class="c0">+</span></p></td><td class="c2"><p class="c8 c16"><span class="c0"></span></p></td></tr><tr class="c12"><td class="c23"><p class="c8"><span class="c0">5-JW</span></p></td><td class="c6"><p class="c8"><span class="c0">+</span></p></td><td class="c10"><p class="c8"><span class="c0">+</span></p></td><td class="c20"><p class="c8"><span class="c0">X</span></p></td><td class="c2"><p class="c8 c16"><span class="c0"></span></p></td></tr><tr class="c12"><td class="c23"><p class="c8"><span class="c0">2B- JW</span></p></td><td class="c6"><p class="c8"><span class="c0">+ (24)</span></p></td><td class="c10"><p class="c8"><span class="c0">X</span></p></td><td class="c20"><p class="c8"><span class="c0">X</span></p></td><td class="c2"><p class="c8 c16"><span class="c0"></span></p></td></tr></tbody></table><ul class="c1 lst-kix_fc43b3h5zds-0"><li class="c5"><span>The transformations with DNA from the well (B2 and P2) had lower efficiencies than those with DNA from a mini-prep. Most likely this is due to drastic differences in DNA concentration (p110+RBS (2) 4/23 was at 254.4ng/ul)</span></li><li class="c5"><span>Conclusions</span></li></ul><ul class="c1 lst-kix_fc43b3h5zds-1 start"><li class="c7"><span>None of the competent cells were contaminated</span></li><li class="c7"><span>All of the competent cells are in fact, competent</span></li></ul><ul class="c1 lst-kix_fc43b3h5zds-0"><li class="c5"><span>Set up O/N of DH5-alpha cells to make competent tomorrow</span></li><li class="c5"><span>Planning ahead</span></li></ul><ul class="c1 lst-kix_fc43b3h5zds-1 start"><li class="c7"><span>Wednesday</span></li></ul><ul class="c1 lst-kix_fc43b3h5zds-2 start"><li class="c15"><span>Make chemically competent DH5alpha cells</span></li><li class="c15"><span>Test chemically competent cells</span></li><li class="c15"><span>Start phage DNA isolation protocol</span></li></ul><ul class="c1 lst-kix_fc43b3h5zds-1"><li class="c7"><span>Thursday</span></li></ul><ul class="c1 lst-kix_fc43b3h5zds-2 start"><li class="c15"><span>If transformation worked, transform cas9 into cells</span></li><li class="c15"><span>Finish phage DNA isolation protocol</span></li><li class="c15"><span>If primers come in, PCR on phage DNA (digestion method)</span></li></ul><ul class="c1 lst-kix_fc43b3h5zds-3 start"><li class="c17 c26"><span>Gibson?</span></li></ul><ul class="c1 lst-kix_fc43b3h5zds-1"><li class="c7"><span>Friday</span></li></ul><ul class="c1 lst-kix_fc43b3h5zds-2 start"><li class="c15"><span>Digest PCR</span></li></ul><ul class="c1 lst-kix_fc43b3h5zds-3 start"><li class="c17 c26"><span>Also pSB1C3 backbone</span></li></ul><ul class="c1 lst-kix_fc43b3h5zds-2"><li class="c15"><span>Gel extraction</span></li><li class="c15"><span>Ligation</span></li></ul><ul class="c1 lst-kix_fc43b3h5zds-1"><li class="c7"><span>Saturday</span></li></ul><ul class="c1 lst-kix_fc43b3h5zds-2 start"><li class="c15"><span>Transformation</span></li></ul><p class="c17 c16"><span></span></p><p class="c17"><span>6/4</span></p><ul class="c1 lst-kix_cgofyvgt0m48-0 start"><li class="c5"><span>Received cas9 from distribution kit (from GOLD): 4M</span></li><li class="c5"><span>Make chemically competent DH5alpha cells</span></li></ul><ul class="c1 lst-kix_cgofyvgt0m48-1 start"><li class="c7"><span>Labeled: 5a (that’s an alpha sign) 6/4 80 or 120ul</span></li></ul><ul class="c1 lst-kix_cgofyvgt0m48-0"><li class="c5"><span>Made chemically competent BW23115 with F-episome</span></li></ul><ul class="c1 lst-kix_cgofyvgt0m48-1 start"><li class="c7"><span>labeled: BWF+ 6/4</span></li><li class="c7"><span>80ul per tube</span></li></ul><ul class="c1 lst-kix_cgofyvgt0m48-0"><li class="c5"><span>Transformation into new competent 5a (6/4)</span></li></ul><ul class="c1 lst-kix_cgofyvgt0m48-1 start"><li class="c7"><span>DNA</span></li></ul><ul class="c1 lst-kix_cgofyvgt0m48-2 start"><li class="c15"><span>p11+RBS(2) (4/23) 1ul</span></li><li class="c15"><span>2B 2ul</span></li><li class="c15"><span>2P 2ul</span></li><li class="c15"><span>2P 3ul</span></li><li class="c15"><span>No DNA</span></li></ul><ul class="c1 lst-kix_cgofyvgt0m48-1"><li class="c7"><span>Plated onto Chlor</span></li><li class="c7"><span>Also, thawed 5a cells for 45 minutes then returned to freezer</span></li></ul><ul class="c1 lst-kix_cgofyvgt0m48-2 start"><li class="c15"><span>Tomorrow, we will transform and test competency</span></li><li class="c15"><span>Also include non-competent control</span></li></ul><ul class="c1 lst-kix_cgofyvgt0m48-0"><li class="c5"><span>Isolation of single-stranded phagemid DNA using M13K07</span></li></ul><ul class="c1 lst-kix_cgofyvgt0m48-1 start"><li class="c7"><span>Added ER2738 colony to 50mL LB</span></li></ul><ul class="c1 lst-kix_cgofyvgt0m48-2 start"><li class="c15"><span>Plate was cold. Next time warm plate before pricking</span></li></ul><ul class="c1 lst-kix_cgofyvgt0m48-1"><li class="c7"><span>After 4 hours, OD was at 0.02. Waited 45 minutes and OD was at 0.08 (somehow). Therefore, we infected at OD 0.08</span></li><li class="c7"><span>Had started another culture when we did not think the first was growing. In incubator for about 1 hour. OD was 0.00. We infected anyway because last time it worked.</span></li><li class="c7"><span>Let infection proceed for 60 minutes then added 70ul of Kanamycin</span></li></ul><ul class="c1 lst-kix_cgofyvgt0m48-0"><li class="c5"><span>Primers came in!</span></li></ul><ul class="c1 lst-kix_cgofyvgt0m48-1 start"><li class="c7"><span>Gem001 F & R, Gem002 F & R, Gem003 F & R</span></li><li class="c7"><span>Resuspended DNA and diluted 1:10</span></li><li class="c7"><span>Other primers were ordered today</span></li></ul><ul class="c1 lst-kix_cgofyvgt0m48-0"><li class="c5"><span>Made assembly of plates with various antibiotic combinations</span></li></ul><p class="c17 c16"><span></span></p><p class="c17"><span>6/5</span></p><ul class="c1 lst-kix_ltqcnt2n1g5e-0 start"><li class="c5"><span>Transformation results from 6/4</span></li></ul><ul class="c1 lst-kix_ltqcnt2n1g5e-1 start"><li class="c7"><span>No growth on negative (no DNA) control: no contamination</span></li><li class="c7"><span>> 400 colonies for positive (p11+RBS (2), diluted 1:10) control. Lawn when not diluted</span></li><li class="c7"><span>~100 colonies for 2B (2ul) not diluted, 18 colonies on 1:10 dilution</span></li><li class="c7"><span>3 colonies for 2P (2ul) not diluted, 0 on 1:10 dilution</span></li><li class="c7"><span>20 colonies for 2P (3ul) not diluted, 1 on 1:10 dilution</span></li></ul><ul class="c1 lst-kix_ltqcnt2n1g5e-0"><li class="c5"><span>Made overnights of 2B and 2P to make freeze downs tomorrow</span></li><li class="c5"><span>Isolated single-stranded M13K07 DNA</span></li></ul><ul class="c1 lst-kix_ltqcnt2n1g5e-1 start"><li class="c7"><span>Final concentration = 5724 ng/ul</span></li></ul><ul class="c1 lst-kix_ltqcnt2n1g5e-2 start"><li class="c15"><span>Calculated from 1:10 dilution which had concentration of 572.4ng/ul</span></li></ul><ul class="c1 lst-kix_ltqcnt2n1g5e-1"><li class="c7"><span>For second sample in pair, we resuspended it in TE but did not proceed to DNA extraction steps</span></li><li class="c7"><span>For the second culture we started 6/4, we resuspended pellet in TBS and glycerol to preserve the M13 phage. Measured absorbances (before glycerol was added)</span></li></ul><ul class="c1 lst-kix_ltqcnt2n1g5e-2 start"><li class="c15"><span>#1</span></li></ul><ul class="c1 lst-kix_ltqcnt2n1g5e-3 start"><li class="c17 c26"><span>269 => 1.690A</span></li><li class="c17 c26"><span>320 => 0.103A</span></li></ul><ul class="c1 lst-kix_ltqcnt2n1g5e-2"><li class="c15"><span>#2</span></li></ul><ul class="c1 lst-kix_ltqcnt2n1g5e-3 start"><li class="c17 c26"><span>269 => 1.453</span></li><li class="c17 c26"><span>320 => 0.059</span></li></ul><ul class="c1 lst-kix_ltqcnt2n1g5e-0"><li class="c5"><span>To biobrick M13ori through biobrick assembly (the old-school way)</span></li></ul><ul class="c1 lst-kix_ltqcnt2n1g5e-1 start"><li class="c7"><span>PCR on Litmus 28i to amplify/biobrick M13ori</span></li></ul><ul class="c1 lst-kix_ltqcnt2n1g5e-2 start"><li class="c15"><span>Used primers Gem003 F & R</span></li><li class="c15"><span>Diluted Litmus 28i DNA 1:10</span></li></ul><ul class="c1 lst-kix_ltqcnt2n1g5e-1"><li class="c7"><span>Digestion of p11+RBS (1) to digest pSB1C3 bb with EcoRI-HF and PstI-HF</span></li><li class="c7"><span>Ran samples on gel and gel extracted pieces. We recieved very low yields (out of range for nano drop)</span></li></ul><ul class="c1 lst-kix_ltqcnt2n1g5e-2 start"><li class="c15"><span>M13 ori: 4.0 ng/ul</span></li><li class="c15"><span>pSB1C3: 1.8 ng/ul</span></li></ul><ul class="c1 lst-kix_ltqcnt2n1g5e-1"><li class="c7"><span>Digested M13 ori fragment despite poor extraction yield with EcoRI-HF and PstI-HF</span></li></ul><ul class="c1 lst-kix_ltqcnt2n1g5e-2 start"><li class="c15"><span>Used 1.5x as much DNA as instructed based on inaccurate concentration</span></li></ul><ul class="c1 lst-kix_ltqcnt2n1g5e-1"><li class="c7"><span>Ligation</span></li></ul><ul class="c1 lst-kix_ltqcnt2n1g5e-2 start"><li class="c15"><span>10hr @ 16C, 10min @ 65C, 4ever @ 4C</span></li></ul><ul class="c1 lst-kix_ltqcnt2n1g5e-0"><li class="c5"><span>To biobrick M13 ori through Gibson Assembly (the cool-kids way)</span></li></ul><ul class="c1 lst-kix_ltqcnt2n1g5e-1 start"><li class="c7"><span>PCR on Litmus 28i</span></li></ul><ul class="c1 lst-kix_ltqcnt2n1g5e-2 start"><li class="c15"><span>Used primers Gem002 F & R</span></li><li class="c15"><span>Diluted Litmus 28i DNA 1:10</span></li></ul><ul class="c1 lst-kix_ltqcnt2n1g5e-1"><li class="c7"><span>PCR on pSB1C3 (p11+RBS (1))</span></li></ul><ul class="c1 lst-kix_ltqcnt2n1g5e-2 start"><li class="c15"><span>Used primers Gem001 F & R</span></li><li class="c15"><span>Diluted pSB1C3 DNA 1:3</span></li></ul><p class="c17 c16"><span></span></p><p class="c17 c16"><span></span></p><p class="c17"><span>6/6</span></p><ul class="c1 lst-kix_bincaufrrv22-0 start"><li class="c5"><span>Ran gel of PCR products from (6/5)</span></li></ul><ul class="c1 lst-kix_bincaufrrv22-1 start"><li class="c7"><span>Recieved bands for pSB1C3 around 2000bp and M13ori around 500bp</span></li><li class="c7"><span>No contamination in pSB1C3 PCR negative control</span></li><li class="c7"><span>Band in M13ori negative control that is the same size as sample. Contaminated by sample?</span></li></ul><ul class="c1 lst-kix_bincaufrrv22-0"><li class="c5"><span>Gibson Assembly</span></li></ul><ul class="c1 lst-kix_bincaufrrv22-1 start"><li class="c7"><span>Diluted pSB1C3 and M13ori PCR products 1:10</span></li><li class="c7"><span>Incubated 60min @ 50C</span></li><li class="c7"><span>Also used provided pUC16 as positive control</span></li></ul><ul class="c1 lst-kix_bincaufrrv22-0"><li class="c5"><span>Transformation</span></li></ul><ul class="c1 lst-kix_bincaufrrv22-1 start"><li class="c7"><span>1. p110+RBS Positive control</span></li><li class="c7"><span>2. No DNA Negative control</span></li><li class="c7"><span>3. Cas9 from distribution kit so we can have more</span></li><li class="c7"><span>4. Thaw and refreeze cells Test competency of comp cells after thawed</span></li><li class="c7"><span>5. Not chem comp cells Negative control for the above</span></li><li class="c7"><span>6. Ligation Product</span></li><li class="c7"><span>7. Gibson product</span></li></ul><ul class="c1 lst-kix_bincaufrrv22-2 start"><li class="c15"><span>7.2. Gibson product diluted 1:4</span></li></ul><ul class="c1 lst-kix_bincaufrrv22-1"><li class="c7"><span>8. Gibson positive control</span></li></ul><ul class="c1 lst-kix_bincaufrrv22-2 start"><li class="c15"><span>7.2. Gibson positive control diluted 1:4</span></li></ul><ul class="c1 lst-kix_bincaufrrv22-1"><li class="c7"><span>For the Gibson product and the positive control, we transformed 2ul of product and 2ul of 1:4 diluted product. NEB recomends the first if using their competent cells and the second if using cells from other companies. Our cells are from NEB but we made them competent ourselves so we tried both ways</span></li></ul><ul class="c1 lst-kix_bincaufrrv22-2 start"><li class="c15"><span>Plated on Chlor at concentrations of 170, 85, and 33 ug/mL</span></li></ul><ul class="c1 lst-kix_bincaufrrv22-0"><li class="c5"><span>Primers came in</span></li></ul><ul class="c1 lst-kix_bincaufrrv22-1 start"><li class="c7"><span>Resuspended and made 1:10 dilutions</span></li></ul><p class="c16 c17"><span></span></p><p class="c17"><span>6/7</span></p><ul class="c1 lst-kix_kjiju65kns7d-0 start"><li class="c5"><span>Results from 6/6 transformation</span></li></ul><ul class="c1 lst-kix_kjiju65kns7d-1 start"><li class="c7"><span>1. Positive control</span></li></ul><ul class="c1 lst-kix_kjiju65kns7d-2 start"><li class="c15"><span>Lots of growth, ~300-400 on 1:10 dilution</span></li></ul><ul class="c1 lst-kix_kjiju65kns7d-1"><li class="c7"><span>2. No DNA negative control</span></li></ul><ul class="c1 lst-kix_kjiju65kns7d-2 start"><li class="c15"><span>No growth</span></li></ul><ul class="c1 lst-kix_kjiju65kns7d-1"><li class="c7"><span>3. Cas9 from distribution kit</span></li></ul><ul class="c1 lst-kix_kjiju65kns7d-2 start"><li class="c15"><span>7 potential colonies (some are close to edges through) on non-diluted</span></li></ul><ul class="c1 lst-kix_kjiju65kns7d-1"><li class="c7"><span>4. Thawed then refroze cells</span></li></ul><ul class="c1 lst-kix_kjiju65kns7d-2 start"><li class="c15"><span>Looks like (1)</span></li></ul><ul class="c1 lst-kix_kjiju65kns7d-1"><li class="c7"><span>5. Not chemically competent cells</span></li></ul><ul class="c1 lst-kix_kjiju65kns7d-2 start"><li class="c15"><span>No growth</span></li></ul><ul class="c1 lst-kix_kjiju65kns7d-1"><li class="c7"><span>6. Ligation product</span></li></ul><ul class="c1 lst-kix_kjiju65kns7d-2 start"><li class="c15"><span>13 potential colonies (some are close to edge)</span></li></ul><ul class="c1 lst-kix_kjiju65kns7d-1"><li class="c7"><span>7. Gibson Assembly Product</span></li></ul><ul class="c1 lst-kix_kjiju65kns7d-2 start"><li class="c15"><span>170 -> No colonies</span></li><li class="c15"><span>85 -> No colonies</span></li><li class="c15"><span>33 -> 3 specks</span></li></ul><ul class="c1 lst-kix_kjiju65kns7d-1"><li class="c7"><span>7.2. Gibson Assembly product diluted 1:10</span></li></ul><ul class="c1 lst-kix_kjiju65kns7d-2 start"><li class="c15"><span>170 -> 1 speck</span></li><li class="c15"><span>85 -> 3 colonies</span></li><li class="c15"><span>33 -> 13 colonies</span></li></ul><ul class="c1 lst-kix_kjiju65kns7d-1"><li class="c7"><span>8. Gibson positive control</span></li></ul><ul class="c1 lst-kix_kjiju65kns7d-2 start"><li class="c15"><span>No colonies</span></li></ul><ul class="c1 lst-kix_kjiju65kns7d-1"><li class="c7"><span>8.2. Gibson positive control diluted 1:4</span></li></ul><ul class="c1 lst-kix_kjiju65kns7d-2 start"><li class="c15"><span>No colonies</span></li></ul><ul class="c1 lst-kix_kjiju65kns7d-0"><li class="c5"><span>Made 6mL O/N cultures</span></li></ul><ul class="c1 lst-kix_kjiju65kns7d-1 start"><li class="c7"><span>4 from (3) cas9 plate</span></li><li class="c7"><span>7 from (6) Ligation product</span></li><li class="c7"><span>5 from (7.2 [85]) Diluted Gibson product on 85 ug/mL Chlor</span></li><li class="c7"><span>8 from (7.2 [33]) Diluted Gibson product on 33 ug/mL Chlor</span></li></ul>
| |
| | | | |
| - | <!-- === To here === -->
| + | ===Additional Work and Characterization (BBa_K1445002)=== |
| - | </div>
| + | Part BBa_J04450 and flanking regions to the sequencing primer sites were cloned into the Litmus28i vector to make a biobricked phagemid backbone that can be packaged into M13 phage. |
| - | </html>
| + | To verify functionality, the vector was packaged into phage using the M13K07 helper phagemid. Progeny phage infected ER2738 ''E. coli'' and cells were selected for on ampicillin. |
| - | <html><li class=<html><div id="gold_Sunday_1st_June.html"></div></html>
| + | |
| - | <html>
| + | [[File:UCB-Results07.jpg|600px|thumb|none|Figure 7. Packaging efficiency of recombinant phagemid to helper phagemid packaging. A) Infected cells diluted 1:1000 and grown on 100 ug/mL Ampicillin to select for BBa_K1445002. B) Infected diluted 1:1000 and grown on 50 ug/mL Kanamycin to select for M13K07 helper phagemid. BBa_K1445002 was packaged over M13K07 at a ratio of 136:1.]] |
| - | <div class ="tbnote">
| + | |
| - | <h2>GOLD LAB</h2>
| + | |
| - | <a href="https://2013.igem.org/Team:Paris_Bettencourt/Project/Detect" target="_blank" class="tbnotelogo PSlogo"> ASDF </a>
| + | |
| - | <div style="clear: both;"></div>
| + | |
| - | <h3>Sunday 1<sup>st</sup> June</h3>
| + | |
| - | <p><b><em>
| + | |
| - | <!-- === Modify from here === -->
| + | |
| - | Protocol and Procedures
| + | |
| - | <!-- === To here === -->
| + | |
| - | </br></em></b></p>
| + | |
| - | Lorem ipsum dolor sit amet, consectetur adipiscing elit. Sed adipiscing luctus suscipit. Sed fringilla est quis adipiscing tempus. Sed ultricies ullamcorper massa, nec euismod orci venenatis vel. Maecenas a eros ut enim laoreet sollicitudin. Aenean faucibus lectus libero, ac consequat quam vehicula a. Proin mattis vestibulum libero, vel congue sapien pretium ut. Curabitur id dapibus turpis. Fusce ultricies arcu vel neque tempus dignissim.
| + | |
| - | </br>
| + | |
| - | Cras lacinia ut justo quis molestie. Nunc nec dolor eleifend, sagittis augue et, ornare diam. Suspendisse iaculis ligula nisl, ac suscipit lacus eleifend in. Duis congue eros et quam tempor, id euismod lectus pharetra. Etiam tincidunt nulla nec elit ultricies vehicula. Maecenas in ornare dolor. Cras eu sem quis tellus lacinia convallis. Nulla ornare, lectus vel aliquam varius, elit lectus congue quam, vel imperdiet libero turpis eu est. Maecenas pellentesque turpis libero, sed viverra lacus egestas sit amet.
| + | |
| - | </br>
| + | |
| - | Morbi luctus euismod laoreet. Nullam neque turpis, viverra id odio eget, tristique dignissim arcu. Nullam dictum, enim non eleifend faucibus, dui mi imperdiet erat, nec iaculis leo tellus id tellus. Praesent neque lacus, volutpat quis elit in, dictum porttitor erat. Vivamus interdum scelerisque ligula vitae accumsan. Sed malesuada porta dignissim. Ut eleifend, erat non fringilla tempus, sem est euismod erat, id tempor ligula nulla tristique ante. Etiam ornare elit nisl, a rutrum ante lobortis vel.
| + | |
| - | </br>
| + | |
| - | In hac habitasse platea dictumst. Cras imperdiet lacus et lacinia mollis. In id ante commodo, posuere ipsum nec, elementum felis. Quisque porta ullamcorper nulla, et porttitor odio elementum eget. Donec iaculis felis sapien, eget scelerisque arcu euismod eget. Sed adipiscing imperdiet mi nec rutrum. Vivamus varius eleifend justo, vitae lacinia nibh volutpat sit amet. Quisque malesuada sodales urna ac commodo. Nunc lobortis augue sem, quis ornare magna vulputate vestibulum. Morbi gravida ullamcorper vehicula. Aenean tristique facilisis venenatis. Fusce vel dignissim risus, eu blandit nibh. In hac habitasse platea dictumst.
| + | |
| - | </br>
| + | |
| - | Donec a blandit mi. Morbi viverra tristique magna at blandit. Etiam vestibulum eu augue eget tincidunt. Pellentesque habitant morbi tristique senectus et netus et malesuada fames ac turpis egestas. Vivamus egestas auctor nibh. Suspendisse sodales ac urna in pellentesque. Sed suscipit egestas justo a dictum. Mauris quis ligula congue, aliquet metus vel, suscipit lorem. Morbi ut viverra sapien. Nulla imperdiet pellentesque purus id dapibus. Suspendisse facilisis auctor arcu nec gravida. Morbi ut euismod lorem. Donec aliquet euismod euismod. Vestibulum facilisis suscipit nisi at posuere. Pellentesque mauris augue, vulputate nec suscipit congue, commodo pharetra dui.
| + | |
| - | <!-- === Modify from here === -->
| + | |
| - | <br>
| + | |
| - | <!-- === To here === -->
| + | |
| - | </div>
| + | |
| - | </html>
| + | |
| | | | |
| | + | |
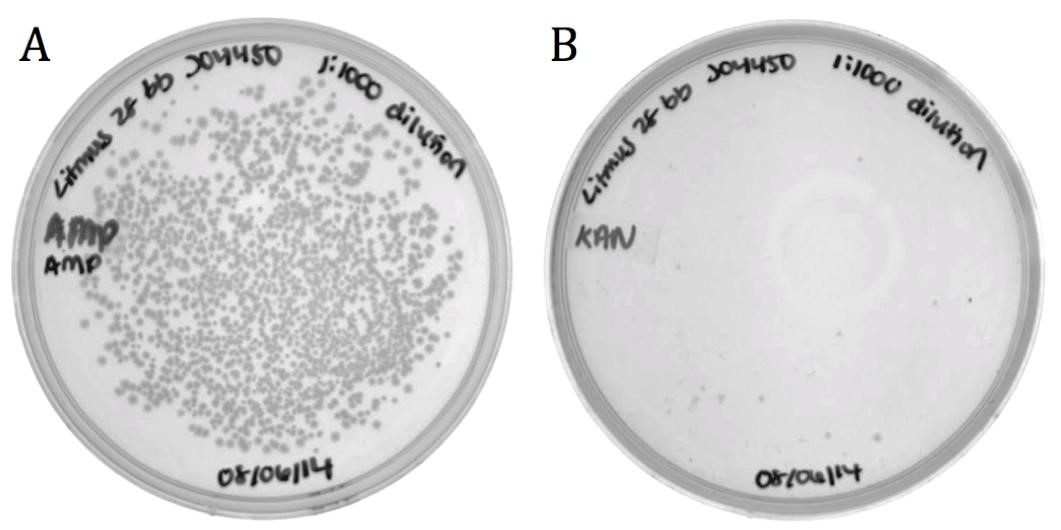
| | + | The growth on ampicillin (1496 colonies) shows that the biobricked version of Litmus28i retains its ability to be packaged and delivered by M13 phage. Additionally, the same sample was plated on kanamycin to determine the packaging ratio of phagemid to helper phagemid. As shown in Figure 7B, very little growth was present on kanamycin (11 colonies) confirming that BBa_K1445002 was preferentially packaged over the M13K07 helper phagemid at a ratio of 136:1. |
| | | | |
| - | </section>
| + | ===References=== |
| - | </body>
| + | ::1. Gomaa AA, Klumpe HE, Luo ML, Selle K, Barrangou R, Beisel CL. 2014. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. mBio 5(1):e00928-13. doi:10.1128/mBio.00928-13. |
| - | <html> | + | <br> |
| - | </div>
| + | <br> |
| - | </div>
| + | <br> |
| - | <div style="clear: both;"></div>
| + | <br> |
| - | | + | {{Template:UCB-Footer}} |
| - | <style>
| + | |
| - | #footer-box{
| + | |
| - | display:none;
| + | |
| - | }
| + | |
| - | </style>
| + | |
| - | | + | |
| - | </html>
| + | |
The system is comprised of two parts: the delivery of the CRISPR-Cas9 machinery and the targeting of the endonuclease.
M13 phage were used as the delivery mode in this system. Depicted in Figure 1 is a single bacterium containing a helper phagemid and a CIRSPR-Cas9 phagemid. The phage coat and assembly proteins are expressed from the helper phagemid, who’s packaging signal has been disrupted by the insertion of a selectable marker (kanamycin in M13K07 or ampicillin in M13g6A1, see Figure 2). Due to the disruption of the helper phagemid’s packaging signal, the probability it will be packaged into a phage capsid has been greatly reduced. The CRISPR-Cas phagemid, BBa_K1445001 contains the endogenous CRISPR-Cas9 sequence from Streptococcus pyogenes and a fully functional packaging signal. M13 phage capsids preferentially package BBa_K1445001 during phage assembly. The resulting phage phage containing the BBa_K1445001 phagemid are isolated and can be used to infect a target population of bacteria.
Once the CRISPR-Cas9 phagemid enters the cell, the bacterium will express the Cas9 endonuclease and a guide RNA(gRNA). Guide RNAs are composed of a spacer sequence that binds the DNA, and a handle to which the Cas9 endonuclease binds. The Cas9 protein and the gRNA come together to search DNA within the cell for PAM sites (Protospacer Adjacent Motifs, shown here in orange). The Cas9 will bind to the PAM site allowing the gRNA to anneal to the target sequence (shown here in green). If binding is successful, the Cas9 endonuclease cleaves the DNA resulting in a double stranded break. If the cell is unable to repair the damage itself or does so incorrectly, the cell will die¹.
The M13 origin of replication (M13ori) has been documented as the packaging signal for the M13 phage. When single stranded, the M13ori forms unique hairpins that signal packaging into a phage capsid. To verify its functionality, its packaging efficiency was compared to that of amilCP. The amilCP sequence is roughly equal in length to the M13ori and both parts were cloned onto the pSB1C3 backbone; therefore, the nucleotide sequence is the only distinguishing factor. The helper phagemid M13K07 made phage in host cells containing pSB1C3-M13ori or pSB1C3-amilCP phagemids. ER2738 E. coli were infected by phage from the M13ori or amilCP sample to assess the respective packaging efficiencies.
The growth in the M13ori sample demonstrates that this phagemid was packaged and delivered by the M13 phage. The absence of growth in the amilCP sample proves that without the presence of a packaging signal, a plasmid cannot be taken up by a phage capsid. These results show that the M13ori is necessary and sufficient for phagemid packaging.
In the phage delivery experiment, M13K07 was used as the helper phagemid. This part contains a kanamycin resistance gene, which is similar to the neomycin phosphotransferase gene being targeted by CRISPR-Cas in this experiment. A new resistance marker was needed to be able to screen future phage production. The genes that code for the phage coat proteins were amplified from M13K07 and cloned into the pSB6A1 vector. Phage amplification, using M13g6A1 as the helper phagemid (and no additional phagemid), produced M13 phage packaging M13g6A1. ER2738 cells were infected with the isolated phage and were plated onto ampicillin to verify the viability of phage produced by this helper phagemid.
The presence of colonies in Figure 2 confirm that M13g6A1 is capable of producing viable phage.
For the following Cas9 experiments, a spacer sequence was designed to target the neomycin phosphotransferase gene present in the targeting strain.
This program inputs FASTA files in two sets. In one are the sequences that should be targeted for Cas9 binding while the other set consists of sequences that should not be targeted. It then searches for possible targeting sites that are present in the targeting sequences and absent from the sequences that should not be targeted. The best spacer sequences are then presented to the user.
For this project, the sequence for the neomycin phosphotransferase gene was given to the program as the targeting sequence. The E. coli strains K-12, BL21 and MG1655 genomes were given to the program as sequences to avoid targeting.
One of the output spacers was chosen for use in the following CRISPR-Cas9 experiments.
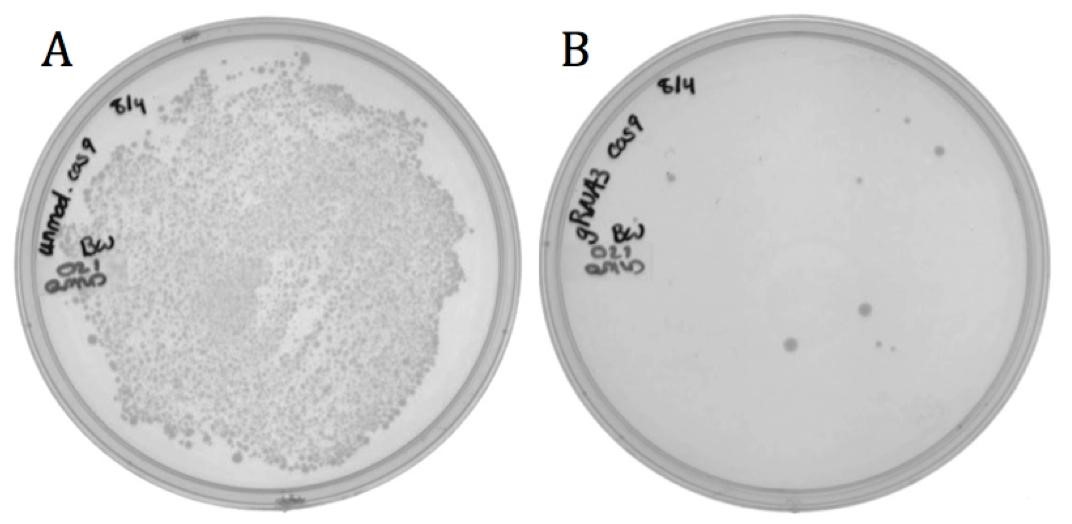
The spacer designed to target the neomycin phosphotransferase gene was cloned into the CRISPR-Cas9 construct (BBa_K1218011). The unmodified spacer was used as a non-targeting control since it does not compliment any region in the BW23115(::kan) or ER2738 genomes.
The decrease in growth between the non-targeting (1920 colonies) and the targeting (8 colonies) samples is credited to the difference in spacer sequence. When the spacer sequence compliments the genome, the Cas9 endonuclease is able to bind and cleave the DNA, resulting in cell death. Interestingly, some targeted colonies survived as can be seen in Figure 5B. Sequencing showed that all eight surviving colonies had deleted the spacer region and one or both of the adjacent repeats. None the less, the difference in transformation efficiency demonstrates the potential for CRISPR-Cas systems as antibacterial therapeutics.
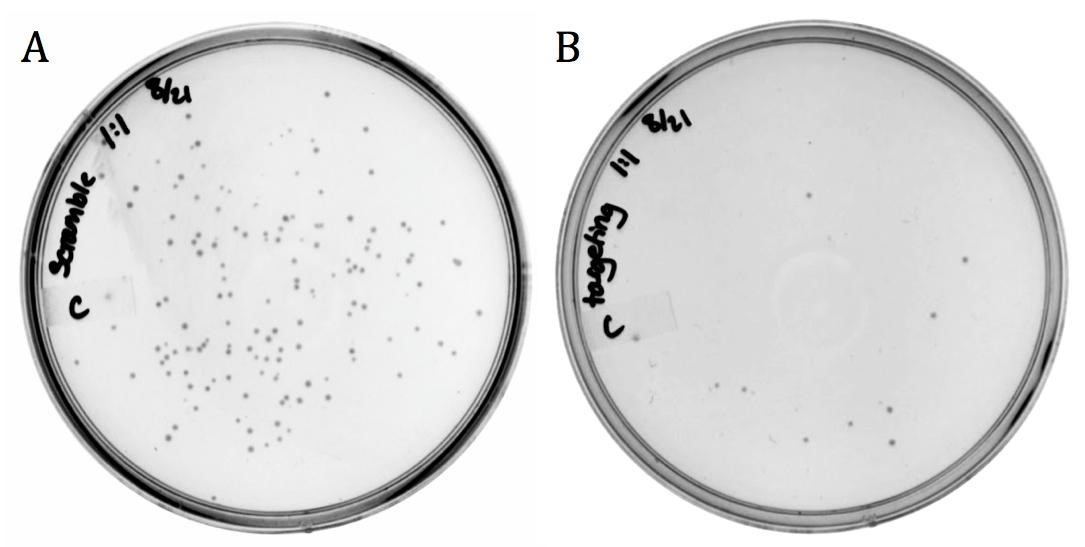
The M13 origin of replication, or M13ori (BBa_K1445000) was cloned upstream of the CRISPR-Cas9 construct (BBa_K1218011) in the targeting and the non-targeting varieties to allow for their uptake into M13 phage. This composite part was packaged into phage using the M13g6A1 helper phagemid. Conjugated BW23115(::kan, F’) E. coli were infected with progeny phage and plated onto chloramphenicol.
The presence of growth in both samples demonstrates that the addition of the M13ori does allow for packaging and delivery by the M13 phage.
The decrease in growth from the non-targeting (143 colonies) to the targeting (11 colonies) is accredited to the differences in spacer sequence. With a spacer sequence that targets within the genome of the cell, the Cas9 endonuclease is able to bind and cleave the DNA, successfully killing the bacterium.
Unmodified spacer may have off-targeted binding to F’ episome, causing death in phage producing cell, thereby reducing phage yield, or in BW23115(::kan, F’) resulting in fewer colonies than expected.
Part BBa_J04450 and flanking regions to the sequencing primer sites were cloned into the Litmus28i vector to make a biobricked phagemid backbone that can be packaged into M13 phage.
To verify functionality, the vector was packaged into phage using the M13K07 helper phagemid. Progeny phage infected ER2738 E. coli and cells were selected for on ampicillin.
 "
"