Team:CU-Boulder/Notebook/Phage Team
From 2014.igem.org
Phage Delivery
Contents |
Week 1
Notes: Unless stated otherwise, all gels contain 2-log ladder
5/9
- Obtained BW23115 KanR cells- BW23115 cells that had their native CRISPR-Cas system knocked out by the insertion of a Kanamycin resistance gene
- -Will also be called BW23115 or BW
- -Conjugated BW23115 KanR cells with contain F’ notation (ex. BWF’)
- Obtained ER2738 cells that contain the F’ episome (no changes from NEB sample)
- -Will also be called ER. Assume that all ER samples contain the F’ episome
- -Streaked sample onto LB+Tet (20ug/mL) to select for colonies containing F’ episome
5/10
- Did receive colonies from 5/9 selection
Week2
5/12
- Need to conjugate BW23115 KanR cells with the F’ episome
- -Set up overnight cultures of ER2738 and BW23115 KanR
- -When mixed, ER2738 will donate it’s F’ episome and BW23115 KanR will receive the F’ episome. F’ episome confers Tetracycline resistance
5/13
- Started M13 Amplification: Amplify M13 phage using the M13K07 Helper Phage
- -Let precipitated in NaCl/PEG solution overnight
- -Possible sources of error
- Did not sterilize 2.5M NaCl/20% PEG-8000 solution
- Added 4-fold PEG solution
- Compensated by adding more LB
- During precipitation, put sample in -20C for 30 minutes before realizing mistake and moving to it to 4C. Sample partially froze
- Conjugated BW23115 with F’ episome
- -Added 1mL BW23115 to 1mL ER2738 overnight culture
- -Incubated at 37C for 30 minutes, shaking
- -Plated on LB+Kan(50ug/mL)+Tet(20ug/mL)
- To select for BW cells that took the F’ episome (containing Tet resistance)
5/14
- Finished the M13 Amplification
- -Visualized product on UV-vis. There was a tall spike at 269nm indicating that DNA was present. Did not test at 320nm.
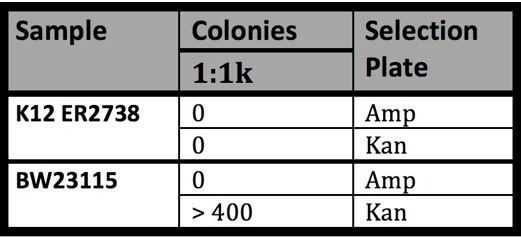
- Results of BW23115 Conjugation
- -Many colonies indicating successful conjugation of F’ episome into BW23115
- -Set up overnight to make freeze down tomorrow
- Set up overnight of ER2738 to make chemically competent tomorrow
5/15
- Made freeze down of BW23115 KanR F’
- -BW23115 E. coli strain with Kanamycin resistance gene inserted into genome and with F’ episome
- Made chemically competent ER2738 cells
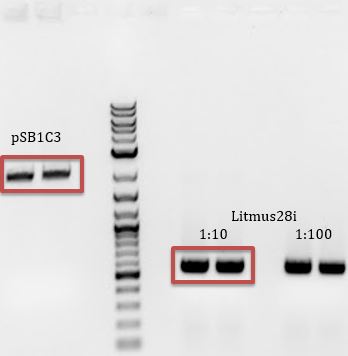
- Transformation of Litmus28i (from NEB) into chemically competent ER2738 cells
- -Added 1ul Litmus28i plasmid to 40ul competent cells
- -Plated on LB + Amp(100ug/mL)
- -Purpose: To make M13 phage that package Litmus28i DNA. Need phagemid (Litmus28i) DNA in infectable cells (cells containing F’ episome) to introduce M13K07 Helper Phage and make phage.
5/16
- Results of 5/15 transformation
- -No growth for No DNA control
- -Many colonies for sample
Week 3
5/19
- M13 Amplification to isolate M13-Litmus28i phage
- -Cells: ER2738 cells containing Litmus28i phagemid
- -Helper Phage: M13K07
- -Not much phage was precipitated
- Set up overnight culture of ER2738 to infect tomorrow
5/20
- Infected ER2738 cells with M13-Litmus28i phage
- -Plated only on Ampicillin(100ug/mL) (should have also plated on kanamycin)
- -Infected for 4-5 hours-> should have only infected for 30 minutes maximum. This extra time gives the cells that were infected with M13-M13K07 the time to produced M13-M13K07 phage and reinfect
5/21
- Results from M13-Litmus28i infection of ER2738
- -Solid lawn of growth for diluted and non-diluted
- -Also sickly looking growth
- Set up overnights
- -ER2738 cells containing Litmus28i for freeze down
- -BW23115 with F’ episome to make chemically competent cells
- -ER2738 to redo infection
5/22
- Tested absorbance of phage produced through M13 amplification on 5/19
- -Low absorbance of 0.018 at 269nm but no detection at wavelength 320nm
- -Decided to redo M13 amplification
- Made chemically competent BW23115 with f-episome
- Made freeze down of ER2738 containing Litmus28i
- Set up overnight of ER2738 containing Litmus28i to redo M13 amplification tomorrow
5/23
- Protocol switch to make phage using phagemid
- -“M13 Amplification” protocol should only be used to make more M13-M13K07, not to make M13 phage containing a different phagemid
- -Switched to new protocol (“Use of M13K07 Helper Phage for isolation of single stranded phagemid DNA” by NEB. Made modifications (see our protocols) to isolate phage rather than single-stranded DNA)
- -Making phage….
| Helper phage | Phagemid | Cells | Notes |
|---|---|---|---|
| M13K07 | None | ER2738 | Make more M13-M13K07 |
| M13K07 | Litmus28i | ER2738 | Test packaging of Litmus28i |
- Made fresh antibiotics
- -Chloramphenicol (34 ng/mL)
- 1.44g chloramphenicol into 42mL EtOH
- -Ampicillin (50 ng/mL)
- 4g ampicillin into 80mL mili-Q H2O
- -Chloramphenicol (34 ng/mL)
5/24
- Isolated phage using new protocol
- -Resuspended pellet in 200ul TBS and 200ul 30% glycerol
- -Measured absorbance with UV-vis
- concentration (phage/mL) = 6x10^16 x (A269-A320)/ (#of base pairs in the phage genome)
| Abs (269nm) | Abs (320nm) | Genome size | Concentration (phage/mL) | |
|---|---|---|---|---|
| M13-M13K07 | 0.721 | 0.060 | 4.57 x10^12 | |
| M13-Litmus28i | 0.250 | 0.028 | 2823 | 4.72 x10^12 |
- Infect ER2738 cells with M13-Litmus28i
- -Wanted 1:10 phage:cell ratio. Math….
- At 1 OD (e.coli), cell/mL = 5x10^8
- 5x10^7 phage * (1mL/4.72x10^12 phage) = 0.011ul phage
- -Wanted 1:10 phage:cell ratio. Math….
- Set up overnights
- -ER2738 for infection with M13-Litmus28i
- -BW23115 F’ for infection with M13-Litmus28i to test infectivity of conjugated strain
Week 4
5/25
- Infect ER and BWF’ cells with M13-Litmus28i
- Made 5mL culture of ER and BW that was at 1 OD
| Sample | OD | mL sample for 1OD in 5mL | mL LB to 5mL |
|---|---|---|---|
| ER2738 | 2.5 | 2 mL | 3 mL |
| BW23115 | 2.0 | 2.5 mL | 2.5 mL |
- -Based on calculations from 5/24, we needed to add 0.011 ul phage per 1 mL of cells at 1 OD. This equates to 0.055 ul of phage into 5 mL cells; therefore we made a 1:10 dilution so we could add 0.5ul. Unfortunately, the pipet would not take up 0.5ul so we added 0.8ul of M13-Litmus28i phage
- -Grew the cells for 20 minutes at 37C
- -Plated 300ul onto Kanamycin (50ug/mL) and 300ul onto Ampicillin (100ug/mL) for each sample
- Incubated overnight at 37C
- Note: During the production of phage, the phagemid SHOULD be packaged preferentially over the Helper Phagemid but some Helper Phagemid will still be packaged. We plated on Amp to select for cells that were infected with phage containing Phagemid. We plated on Kan to select for cells that were infected with phage containing Helper Phagemid. This allows us to compare the packaging efficiency of Helper Phagemid: Phagemid.
5/26
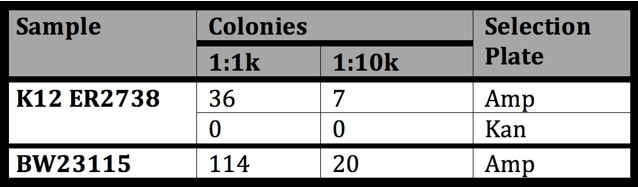
- Results from 5/25 infection with M13-Litmus28i
| Sample | Result | Significance |
|---|---|---|
| ER2738 on Amp | Lawn | Litmus28i phagemid was successfully packaged into the M13 phage and is infectable |
| ER2738 on Kan | 100-200 colonies | Some M13 helper phage is packaged into the M13 phage but at a much lower rate than Litmus28i |
| BW23115 on Amp | Lawn | BW23115 is ‘equally’ infectable by M13 as ER2738 |
| BW23115 on Kan | Lawn | BW23115 contains Kan resistance in its genome so this tells us nothing |
- Conclusions:
- -Cells grew on Ampicillin; therefore, Litmus28i phagemid was successfully packaged into M13 phage.
- -For ER2738 samples, there was significant growth on Ampicillin compared to Kanamycin; therefore, Litmus28i phagemid is packaged preferentially over M13K07 Helper Phagemid
- -M13-Litmus28i retains its infectivity of cells containing the F’ episome
- Because we received lawns, we have to redo the infection and plate less cells so we can calculate the uptake ratio between the phagemid and helper phage based on the number of colonies
- Started 50 mL overnight of K12 ER2738 and BW23115
5/27
- Redo the infection done on 5/25
- -Infectable cells: ER2738 and BW23115
- Plated non-infected samples of each (non-diluted) to check for contaminants
- -Diluted M13-Litmus28i (1) phage by a factor of 10. Added 5.5ul to each sample
- -Grew samples for 20 minutes at 37C, 250rpm
- -Plated 100ul of onto an Ampicillin (100ug/mL) plate and onto a Kanamycin (50ug/mL) plate. Incubated overnight at 37C.
- Dilutions = 1:10; 1:100; and 1:1000
- -Infectable cells: ER2738 and BW23115
5/28
- Results from 5/27
- -Controls were as expected
- No growth for ER2738 non-infected grown on Amp, ER2738 non-infected grown on Kan, or BW23115 non-infected grown on Amp
- Growth for BW23115 non-infected grown on Kan (BW23115 has Kan R in genome)
- -Many colonies were received for all dilutions (1:10, 1:100, and 1:1000) of the following
- ER2738 infected and plated on Amp
- BW23115 infected and plated on Amp
- BW23115 infected and plated on Kan
- -Many (100s to 1000?) colonies grew on 1:10 and 1:100 dilutions of ER2738. 50-100 colonies grew on the 1:1000 dilution of ER2738
- Compare this to the 100-200 colonies that grew from 2/25 infection (which was 300ul non-diluted, infected cells)
- Reasons for increased yield
- Added too much phage?
- volume changed between experiment (5mL to 50mL)
- Overnight culture may not have been saturated. If still in log phase, the cells would continue to grow
- -Controls were as expected
- Made 50mL O/N cultures of ER2738 and BW23115 so we can repeat the infection tomorrow and plate further dilutions starting at 1:1000
- -Carry out infection in 5mL and 50mL to test volume effect?
5/29
- Measured OD of overnights
| Sample | OD | mL to have .1OD in 50mL | mL to have 1OD in 5mL |
|---|---|---|---|
| K12 ER2738 | 3.0 | 1.7 | 1.7 |
| BW23115 | 2.9 | 1.7 | 1.7 |
- Experiment 1: Infect cells using same method as 5/25 (in a 5mL culture)
- -Started with 1OD cells in 5mL
- -Added about 0.7ul (inaccuracies in pipet) of 1:10 diluted M13-Litmus28i phage
- -Incubated (rotating) for 20 minutes
- -Made 1:1,000 and 1:10,000 dilutions
- -Plated 100ul on Ampicillin (100ug/mL) plates and on Kanamycin (50ugmL) plates
- Included non-infected samples diluted by 1:1000
- This negative control can be used for Experiment 2 since the non-infected parent solution is the same
- -Incubate overnight at 37C
- Experiment 2: Infect cells using protocol from “Eliminating helper phage from phage display”
- -Diluted O/Ns to OD of 0.1 in 50 mL culture
- -Grew samples until of OD of ER2728 = 0.59 and the OD of BW23115 = 0.60
- Missed OD of 0.5 mark, but the two samples are close to each other
- -Chilled samples on ice for 30 minutes
- -Warmed in incubator for 35 minutes (should have been 30)
- -Amount of phage. Rather than use 1:1 as mentioned in protocol, we used multiplicity of 1:10 (phage:cell)
- Added 3.3ul of 1:10 diluted M13-Litmus28i (1) phage
- (On 5/27 we added 5.5ul of diluted phage to 50mL of cells at OD of 1. Our cells were at OD of .6; therefore, 5.5*.6 = 3.3ul)
- -Incubated for 30 minutes at 37C, not shaking
- We later change this to shaking
- -Dilutions
- 1:1,000; 1:5,000; 1:10,000; 1:50,000; 1:100,000; 1:1,000,000
- Plated ER2738 and BW23115 on Ampicillin (100ug/mL)
- Plated ER2738 on Kanamycin (50ug/mL)
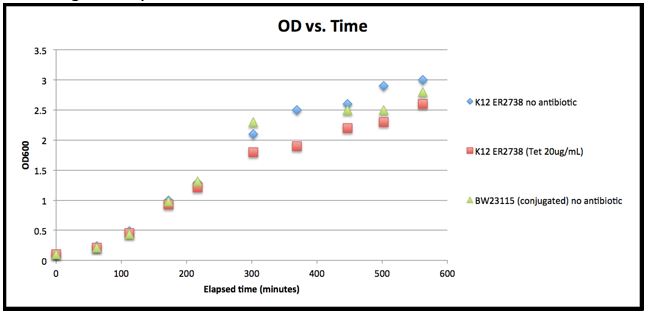
- Experiment 3: Growth Test (for growth curve)
- -We were concerned by the low OD of the Overnights from the last few days. Wanted to be sure that 2.0-3.0 was not still in log phase. Cultures looked saturated but the OD seemed low.
| Time | Elapsed time (min) | ER2738 (no antibiotic) | ER2738 (Tetracycline (20ug/mL)) | BW23115 (with F’ episome) no antibiotic |
|---|---|---|---|---|
| 10:08 | 0 | 0.1 | 0.1 | 0.1 |
| 11:10 | 62 | 0.24 | 0.21 | 0.21 |
| 12:00 | 112 | 0.49 | 0.45 | 0.44 |
| 13:00 | 172 | 1.00 | 0.93 | 0.98 |
| 14:15 | 217 | 1.29 | 1.21 | 1.31 |
| 15:40 | 302 | 2.1 | 1.8 | 2.3 |
| 16:47 | 369 | 2.5 | 1.9 | 3.4 |
| 18:05 | 447 | 2.6 | 2.2 | 2.5 |
| 19:00 | 502 | 2.9 | 2.3 | 2.5 |
| 20:00 | 562 | 3.0 | 2.6 | 2.8 |
- The time point at 16:47 (369 minutes elapsed) for BW23115 conjugated (without antibiotics) is most likely an error. It has been removed from the growth plot
- Other
- -Made Amp and Kan plates (1 sleeve of each)
- -Made 50mL O/N of ER2738 and BW23115F’ in case we need further dilutions
- -Made 5mL O/N of ER2738, BW23115F’, and BW23115 (without F’ episome) to make chemically competent tomorrow
- Did not have plate of BW23115 (without F’ episome) so used freeze down. Hoping to get O/N of a picked colony from CRISPR Team tomorrow morning
5/30
- Made chemically competent cells of…
- -ER2738
- -BW23115F’ (conjugated with F’ episome)
- -BW23115 (not conjugated- without F’ episome)
- Culture started from plate
- -BW23115* not conjugated (without F’ episome)
- Culture started from freeze down
- Results from infections
- -Negative Controls (cells were not infected; cells were diluted 1:1000)
- -Results from Experiment 1 (5/29)
- -Results from Experiment 2 (5/29)
- Math
- -If there are 5.00E+8 cells in 1mL of culture at OD of 1, then in 1mL of culture at OD of 0.59, there are 2.95E+8 cells. In a 50mL culture at OD of 0.59, there are 1.48E+10 cells.
- -We added 3.3ul (0.0033mL) phage at concentration 4.62E+11 phage/mL which amounts to 1.52E+9 total phage
- -Assuming that 1 phage infects 1 bacterium, we can assume that 1.52E+9 bacterial have the potential to be infected in the 50mL culture
- -We plated 100ul of culture at various dilutions. If not diluted, the number of cells that can be potentially infected in 0.1mL equals 1.52E+9/500, or 3.05E+06 cells. We then accounted for the dilutions (for 1:1000 dilution, we divided 3.05E+06 by 1000 to receive 3.05E+03)
- -The following table contains the number of cells with the potential to be infected assuming a 100% infectivity rate by M13 phage and that 1 cell is infected only once.
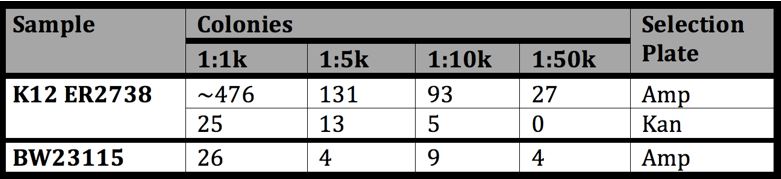
| Dilution | Potential infected cells | Colonies on Amp | % Potential (Amp) | Colonies on Kan | % Potential(Kan) | Kan:Amp |
|---|---|---|---|---|---|---|
| 1:1000 | 1.52E+06 | 476 | 15.62% | 15 | 0.820% | 1:19.04 |
| 1:5000 | 3.05E+05 | 131 | 21.49% | 13 | 2.133% | 1:10.08 |
| 1:10000 | 1.52E+05 | 93 | 30.51% | 5 | 1.640% | 1:18.60 |
| 1:50000 | 3.05E+04 | 17 | 44.29% | 0 | 0.000% |
- Conclusions from infections
- -Results between and within the three trials are inconsistent. For example, the number of colonies received in experiments 1 and 2 from 5/29 differ greatly. Due to the differences in protocol, variation was expected but not to this extent.
- -Our dilutions did not yield the expected 10 fold (or 5 fold) decrease in growth that was expected.
- -Plates from 5/29 could be plated better to reduce dense areas of growth and growth around the rim.
- -Though the experiment contained many errors we can say that the phagemid (Litmus 28i) is preferentially packaged compared to the helper phage (M13K07) but not to the degree we expected.
- -Could receive increased occurrences of cells containing M13k07 due to infection, phage production, further infection
Week 5
6/2
- Tested chemically competent cells through transformation
- -Are cells contaminated?
- -Are cells competent?
- The samples for transformation
| # | Cells (Tube label) | DNA (Tube label) | Resistance before transformation | Resistance after transformation |
|---|---|---|---|---|
| 1 | K12 ER2738 5/20 | p110+RBS (2) 4/23 | Tet | Tet, Chlor |
| 2 | BW (-f) 5/30 | p110+RBS (2) 4/23 | Kan | Kan, Chlor |
| 3 | BW f-ep comp 5/22 | p110+RBS (2) 4/23 | Kan, Tet | Kan, Tet, Chlor |
| 4 | BW (+f) 5/30 | p110+RBS (2) 4/23 | Kan, Tet | Kan, Tet, Chlor |
| 5 | *BW23115 5/30 | p110+RBS (2) 4/23 | Kan | Kan, Chlor |
| 2B | K12 ER2738 | 2B [from dis. kit] | Tet | Tet, Chlor |
| 2P | BW f-ep comp 5/22 | 2P [from dis. kit] | Kan, Tet | Kan, Tet, Chlor |
6/3
- Results from 6/2 Transformation
| Sample | Growth on Chlor | Growth on Kan | Growth on Kan+Tet | Growth on Amp |
|---|---|---|---|---|
| 1 N | X | X | X | X |
| 2 N | X | + | X | X |
| 3 N | X | + | + | X |
| 4 N | X | + | + | X |
| 5 N | X | + | X | X |
| 1 | + | X | X | |
| 2 | + | + | X | |
| 3 | + | + | + | |
| 4 | + | + | + | |
| 5 | + | + | Many colonies but close to samp. 4 | |
| 2P | + (~100) | + | + | |
| 1- JW | + | X | X | |
| 2- JW | + | + | X | |
| 3- JW | + | + | + | |
| 4- JW | + | + | + | |
| 5-JW | + | + | X | |
| 2B- JW | + (24) | X | X |
- The transformations with DNA from the well (B2 and P2) had lower efficiencies than those with DNA from a mini-prep. Most likely this is due to the differences in DNA concentration (p110+RBS (2) 4/23 was at 254.4ng/ul)
- Conclusions
- -None of the competent cells were contaminated
- -All of the competent cells are in fact, competent
- Set up O/N of DH5-alpha cells to make competent tomorrow
6/4
- Isolation of single-stranded phagemid DNA using M13K07
- -Added ER2738 colony to 50mL LB
- Plate was cold. Next time warm plate before pricking
- Best to use freshly grown plate
- -After 4 hours, OD was at 0.02. Waited 45 minutes and OD was at 0.08. Therefore, we infected at OD 0.08
- -Had started another culture when we did not think the first was growing. In incubator for about 1 hour. OD was 0.00. We infected anyway because last time it worked.
- -Let infection proceed for 60 minutes then added 70ul of Kanamycin to be a final concentration of 70ug/mL
- -Added ER2738 colony to 50mL LB
- Primers came in to biobrick M13ori (packaging signal on Litmus28i)
- -Resuspended primers and diluted 1:10
6/5
- Isolated single-stranded M13K07 DNA
- -Final concentration = 5724 ng/ul (calculated from a 1:10 dilution)
- -For second sample in pair, we resuspended it in TE but did not proceed to DNA extraction teps
- -For the second culture we started 6/4, we resuspended pellet in TBS and glycerol to preserve the M13 phage. Measured absorbances (before glycerol was added)
- #1
- 269 => 1.690A
- 320 => 0.103A
- #2
- 269 => 1.453
- 320 => 0.059
- For our first biobrick, we wanted to isolate the M13 origin, a segment ~500bp that allows for packaging into the M13 phage. We tried to achieve this by biobrick assembly and by Gibson Assembly.
- -To biobrick M13 ori through biobrick assembly (the old-school way)
- PCR on Litmus 28i to amplify/biobrick M13ori
- Used primers Gem003 F & R
- Diluted Litmus 28i DNA 1:10
- Digestion of p11+RBS (1) to digest pSB1C3 bb with EcoRI-HF and PstI-HF
- Ran samples on gel and gel extracted pieces. We recieved very low yields (out of range for nano drop)
- M13 ori: 4.0 ng/ul
- pSB1C3: 1.8 ng/ul
- Digested M13 ori fragment despite poor extraction yield with EcoRI-HF and PstI-HF
- Used 1.5x as much DNA as instructed based on inaccurate concentration
- PCR on Litmus 28i to amplify/biobrick M13ori
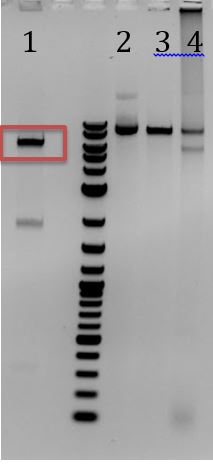
- -To biobrick M13 ori through biobrick assembly (the old-school way)
- -Gel extracted red rectangles
- -Ligation
- 10hr @ 16C, 10min @ 65C, 4ever @ 4C
- To biobrick M13 ori through Gibson Assembly (the cool-kids way)
- -PCR on Litmus 28i
- Used primers Gem002 F & R
- Diluted Litmus 28i DNA 1:10
- -PCR on pSB1C3 (p11+RBS (1))
- Used primers Gem001 F & R
- Diluted pSB1C3 DNA 1:3
- -PCR on Litmus 28i
6/6
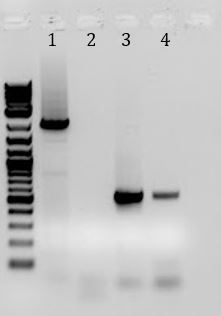
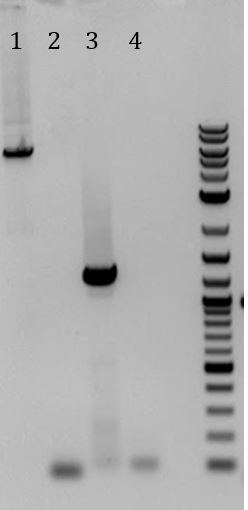
- Ran gel of PCR products from (6/5). Products will be used for Gibson Assembly
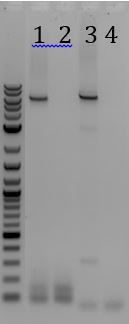
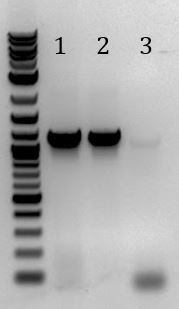
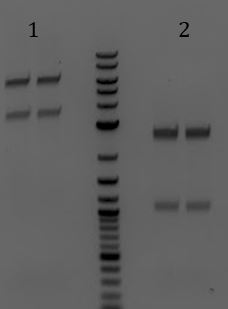
- -Recieved bands for pSB1C3 around 2000bp and M13ori around 500bp
- -No contamination in pSB1C3 PCR negative control
- -Band in M13ori negative control that is the same size as sample. Contaminated by sample?
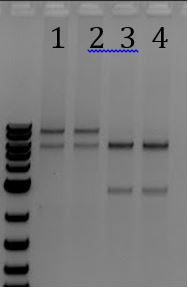
- -Gel of PCR products #1 and #2 from 6/5
- 1. pSB1C3 with promoter+RBS as insert. Amplified with Gem001
- 2. No DNA control for Gem001
- 3. M13ori amplified with Gem002 from Litmus28i
- 4. No DNA control for Gem002
- Gibson Assembly
| Total Amount of Frag. | .02-.5pmol | 10 ul total |
|---|---|---|
| Gibson Assembly MM (2x) | 10 ul | 10 |
| Dionized H2O | 10-x |
- -Diluted pSB1C3 and M13ori PCR products 1:10
- -Incubated 60min @ 50C
- -Also used provided pUC16 as positive control
- Transformation
- 1. p110+RBS Positive control
- 2. No DNA Negative control
- 3. Cas9 from distribution kit so we can have more
- 4. Thaw and refreeze cells Test competency of comp cells after thawed
- 5. Not chem comp cells Negative control for the above
- 6. Ligation Product
- 7. Gibson product
- 7.2. Gibson product diluted 1:4
- 8. Gibson positive control
- 7.2. Gibson positive control diluted 1:4
- -For the Gibson product and the positive control, we transformed 2ul of product and 2ul of 1:4 diluted product. NEB recommends the first if using their competent cells and the second if using cells from other companies. Our cells are from NEB but we made them competent ourselves so we tried both ways
- Plated on Chlor at concentrations of 170, 85, and 33 ug/mL
- Primers came in
- -Resuspended and made 1:10 dilutions
6/7
- Results from 6/6 transformation
- 1. Positive control
- Lots of growth, ~300-400 on 1:10 dilution
- 2. No DNA negative control
- No growth
- 3. Cas9 from distribution kit
- 7 potential colonies (some are close to edges through) on non-diluted
- 4. Thawed then refroze cells
- Looks like (1)
- 5. Not chemically competent cells
- No growth
- 6. Ligation product
- 13 potential colonies (some are close to edge)
- 7. Gibson Assembly Product
- 170 -> No colonies
- 85 -> No colonies
- 33 -> 3 specks
- 7.2. Gibson Assembly product diluted 1:10
- 170 -> 1 speck
- 85 -> 3 colonies
- 33 -> 13 colonies
- 8. Gibson positive control
- No colonies
- 8.2. Gibson positive control diluted 1:4
- No colonies
- Made 6mL O/N cultures
- -4 from (3) cas9 plate
- See Constitutive CRISPR notebook for more info on these samples
- -7 from (6) Ligation product
- -5 from (7.2 [85]) Diluted Gibson product on 85 ug/mL Chlor
- -8 from (7.2 [33]) Diluted Gibson product on 33 ug/mL Chlor
- -4 from (3) cas9 plate
Week 6
6/8
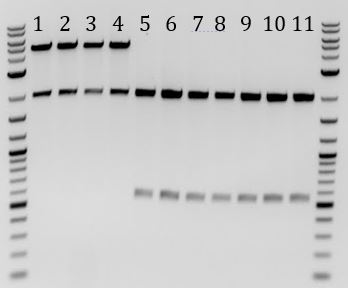
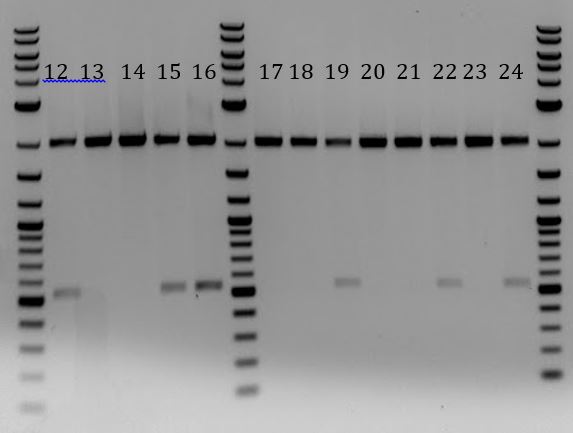
- Check colonies for correct constructs.
- -Mini-prepped all 24 O/Ns
- Yielded low concentrations for samples 12, 15, 19, and 22
- -Digested all with EcoRI and PstI (10ul reactions)
- -Ran results on gel
- All 4 cas9 samples had the expected bands of 2000 and 5000bp
- All 7 ligation products have expected bands of 2000 and ~570bp
- 3 of 5 Gibson assemblies from 85ug/mL Chlor plate had expected bands of 2000 and ~500bp
- 3 of 8 Gibson assemblies from 33ug/mL Chlor plate had expected bands of 2000 and ~500bp
- -Mini-prepped all 24 O/Ns
- 1-4: Cas9 from Stanford-Brown team
- 5-11: pSB1C3-M13ori cloned through ligation
- 12-24: pSB1C3-M13ori cloned through Gibson Assembly
- 12-16: Grown with 85 ug/mL Chlor
- 17-24: Grown with 33 ug/mL Chlor
- Conclusions from gel
- -We have cas9 safely in cells
- -Our ligation reactions successfully yielded M13ori on pSB1C3
- -Combined, we had a 46% success rate for the Gibson Assembly in yielding M13ori on pSB1C3
- The 4 samples that had the lowest concentration after being mini-prepped (12,15, 19, and 22) correlate with samples that had the correct band pattern
- We selected 4 samples from each (4 total between the two Gibson reactions) type
- -For non-Gibson Assembled samples
- Plated 25ul on 170ug/mL Chlor
- -For non-Gibson Assembled samples
| Our usual method | Gibson Method |
|---|---|
| 1. Thaw on ice | 1. Thaw on ice |
| 2. Transfer 40ul cells to tube | 2. Transfer 50ul cell to tube |
| 3. Add DNA. 1ul for mini-prep OR up to 10ul for ligation | 3. Add 2ul to NEB cells OR 2ul of 1:4 diluted to other cells |
| 4. Mix by pipet Let sit 30min on ice | 4. Mix by pipet or flicking Let sit 30 min. on ice |
| 5. Heat shock: 42C for 45s | 5. Heat shock: 42C for 30s |
| 6. Ice for 5 minutes | 6. Ice for 2 min. |
| 7. Transfer to culture tube; Add 200ul SOC | 7. Add 950ul SOC to tube |
| 8. Shake or rotate for 60-120min at 37C | 8. Shake (250rpm) or rotate for 60 min. at 37C |
| 9. | 9. Warm plates to 37C |
| 10. Plate 100ul onto selection plate | 10. Plate 100ul onto plate |
| 11. Incubate O/N @ 37C | 11. Incubate O/N @ 37C |
- -Added 6mL LB and Chlor at concentration of 170ug/mL to grow O/N
- For Gibson Assembled samples
- -Plated 25ul onto 170, 85, and 33ug/mL Chlor
- -Samples from 85ng/mL plate
- Transferred 100ul to new tube, added media, added Chlor at 170ng/mL
- -Samples from 33ng/mL plate
- Tranfered 100ul to new tubes, added media, added Chlor at 85ng/mL to one and 33ng/mL to the other
- Tomorrow, may send for sequencing and make freeze downs
- Transformation
- 1. Positive control (p110+RBS diluted 1:10)
- 2. Non-diluted Gibson product
- 3. Gibson product diluted 1:4
- 4. Gibson product diluted 1:10
- -Transformed each sample using our usual method and using the protocol given by Gibson
- -Due to not have plates ready before transformation, in step 4, the samples sat for about 50 minutes. Then in step 8, they both recovered for about 150 minutes. Though not specified in our protocol, we did warm the plates to 37C. In step 10 for our protocol, since we only added 200ul SOC and wanted to plate on 3 selection plates (see below), we only plated 50ul (except for the positive control).
- -Plated on three concentrations of Chloramphenicol (33 ug/mL, 85 ug/mL, and 170 ug/mL) to determine the differences in yield due to differences in concentration.Obvious hypothesis: more colonies will grow on plates that have a lower concentration of chlor.
6/9
- Made chemically competent 5alpha cells with Dan and Alex from main campus
- -Waiting to hear results on competency
- Will eventually make phage containing CRISPR-Cas9 that targets Kanamycin resistance. M13K07 has Kanamycin resistance so we need to switch the resistance on the M13 genes.
- -PCR on pwp 2.po (plasmid that Sam gave us that contains the zeoR gene adjacent to ori) to amplify zeoR and ori. Zeo is on EM7 promoter
- -Primers: Gem008 R & R
- -Anneal temp from NEBuilder: 63.7C
- -Extension time: 90s
- -Expected band size on gel: 1300bp
- -Used phusion polymerase
- -PCR on pwp 2.po (plasmid that Sam gave us that contains the zeoR gene adjacent to ori) to amplify zeoR and ori. Zeo is on EM7 promoter
- PCR on M13K07 DNA to amplify M13 phage genes (also removes majority of M13 ori, all of KanR, and all of p15 ori)
- -Primers: Gem007 F & R
- -Anneal temp from NEBuilder: 60.2C
- -Extension time: 4:30
- -Expected band size on gel: about 6000bp
- -Used phusion polymerase
- Freeze downs
- -Note: Phagemid 1C3 was the original name for ‘pSB1C3-M13ori’
| Top Label | Side label | What? |
|---|---|---|
| Phagemid 1C3 6/9 | From Lig | M13 ori inserted into 1C3 (biobricked); Done through ligation; Contains extra bases as spacer between biobrick prefrix/suffix and part for primer design |
| Phagemid 1C3 6/9 | From Lig | “ “ |
| Phagemid 1C3 6/9 | Gibson | M13 ori inserted into 1C3 (biobricked); Done through Gibson cloning |
| Phagemid 1C3 6/9 | Gibson | “ “ |
6/10
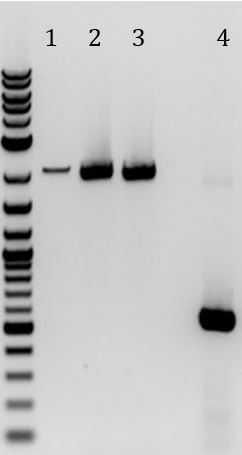
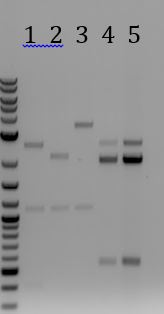
- Ran gel of PCRs from 6/9
- 1. Amplification of M13 genes from M13K07 (~6000bp)
- 2. No DNA control for (1) amplification
- 3. Amplification of Zeo resistance gene + plasmid ori (~1300bp)
- 4. No DNA control for (3) amplification
- Gibson Assembly of above parts (did not gel extract)
- -Diluted the PCR products 1:10 then added 3ul of M13K07 genes product and 7ul of ZeoR+ori product
- -Incubated at 50C for 60 min.
- -Transformed Gibson Assembly product into 5alpha cells
- Used our usual protocol
- Add 2ul of DNA
- -In one sample, diluted DNA 1:4 and in the other, we diluted DNA 1:10
- Started Phage Amplification Protocol
- -ER2738 transformed with Litmus 28i
- Grew for ~2.5hr before reaching an OD of 0.04
- -ER2738 transformed with pSB1C3-M13ori (M13ori on pSB1C3)
- Grew for ~ 3.5hr before reaching an OD of 0.01, then in the next 1.5 hours, spiked to 0.19
- We gave up and went home, and will restart tomorrow
- -ER2738 transformed with Litmus 28i
- Analyzed transformation results from 6/8
6/11
- Made chemically competent 5alpha cells
- Restarted Phage Amplification Protocol
- -Forgot to add phagemid antibiotic at start of growth. Added phagemid antibiotic when we added phage. Incubated for 90 minutes before adding Kanamycin (to select for cells that were infected by M13K07)
- -Phage is at concentration 4.57x10^12 phage/mL
- Protocol calls for final concentration of 1 x10^8 phage/mL
- (4.57 x10^12)*V = (1 x10^8)(50mL)
- V = 0.00109mL
- Added 1.1ul of phage
- Protocol calls for final concentration of 1 x10^8 phage/mL
- Transformations
| DNA | Plate Selection |
|---|---|
| No DNA control | (all) |
| Positive control (p110+RBS) | (C) |
| M13 genes + ZeoR ori | (Z) |
| M13 genes +ZeoR ori diluted 1:4 | (z) |
6/12
- Transformation Results from 6/10 [Took ~36 hours to be clearly visible]
- No DNA control
- -Amp: 0
- -Zeo (50ug/mL): 200 colonies
- M13 genes + ZeoR ori (1:4 dilution)
- -Zeo (25ug/mL): 300
- -Zeo (50ug/mL): 200
- -Zeo (100ug/mL): 100
- M13 genes + ZeoR ori (1:10 dilution)
- -Zeo (25ug/mL): 300
- -Zeo (50ug/mL): 150
- -Zeo (100ug/mL): 150
- No DNA control
- Transformation Results for 6/11
- -Positive (p110+RBS) on Chloramphenicol: 500 colonies
- -No DNA on Amp: 0 colonies Zeo (100 ug/mL): specks
- -No apparent growth on any other plate
- Realized later that we grew our samples on the wrong plates. Will repeat transformation today
- Transformation #1
- This morning there were no colonies on positive (p110+RBS) coltrol from 6/11 even though we observed fast growth in the past. Without waiting for colonies to appear, we started a control transformation
| Sample | Diluted | Time at 42C |
|---|---|---|
| p110+RBS | No | 45s |
| p110+RBS | 1:10 | 45s |
| p110+RBS | 1:10 | 30s |
| p110+RBS | 1:10 | No timer. ~43s |
| No DNA control | No | 45 |
- Because ‘No DNA control 6/10’ yielded colonies, we researched Zeocin plates
- -According to Life Technologies (Invitrogen), Zeocin requires low salt medium and a pH of 7.5
- -Low Salt LB Medium (1L)
- Ingredients
- 10g Tryptone
- 5g NaCl
- 5g Yeast Extract
- -Mix ingredients
- -Adjust pH to 7.5 using NaOH (If go over, use HCl)
- -Add agar for plates at 15g/L. Autoclave
- -Thaw Zeocin on ice. Vortex
- -Add Zeocin to final concentration of 25ug/mL
- Transformation #2
- -Repeat of transformation on 6/11 but this time we will plate on the correct plates
- -Also remade Zeocin plates
- Finished isolation of M13 Litmus phage and M13 pSB1C3-M13ori phage
- Note: Phagemid 1C3 was the original name for ‘pSB1C3-M13ori’
| Phage Sample | A269 | A320 | Concentration (phage/mL) |
|---|---|---|---|
| Litmus phage (1) | 0.181 | 0.034 | 3.12 x10^12 |
| Litmus phage (2) | 0.227 | 0.047 | 3.83 x10^12 |
| Phagemid 1C3 phage (1) | 0.101 | 0.020 | 1.87 x10^12 |
| Phagemid 1C3 phage (2) | 0.126 | 0.021 | 2.42 x10^12 |
- phage/mL = 6x10^16 x (A269-A320)/ (#of base pairs in the phage genome)
- Set up 50mL O/N of K12 ER2738 (containing f-episome) for infection tomorrow with Litmus phage and pSB1C3-M13ori phage
6/13
- Transformation Results for 6/11
- -Several hundred colonies on Positive Control (p110+RBS) on Chlor
- -No colonies for GA positive control on Amp
- -No colonies for M13ori + ZeoR mistakenly plated on Amp
- -No colonies for No DNA control on Amp
- -100-ish colonies for No DNA control on Zeo
- -100-ish colonies for cas9+AmpR+gRNA mistakenly plated on Zeo
- -These last 3 points suggest/confirm that Zeo plates are no good
- Transformation Results from 6/12 control test
- -Colonies grew is about equal amounts on all plates, including No DNA control
- Either plates don’t contain Chlor or competent cells are contaminated
- -Streaked competent cells onto new and old Chlor plates
- -Colonies grew is about equal amounts on all plates, including No DNA control
- Infection test of ER2738: Is Litmus preferentially packaged over M13K07 Helper phage? Is pSB1C3-M13ori preferentially packaged?
- -Infect ER2738 with phage produced 6/12
- Phage should have packaged Litmus 28i phagemid or pSB1C3-M13ori
- Cells infected with phage packaging Litmus 28i will grow on Amp
- Cells infected with phage packaging pSB1C3-M13ori will grow on Chlor
- Cells infected with phage packaging M13K07 will grow on Kan
- -After we plate, we can count the colonies and calculate a ratio of Litmus28i: M13K07 or pSB1C3-M13ori:M13K07 packaging
- -Infect ER2738 with phage produced 6/12
6/14
- Results of contamination test (streaked competent cells onto new and old Chlor plates)
- -Colonies grew in low amounts on both plates. most likely the cells are contaminated
- Transformation results for 6/12
- -Many colonies for No DNA control on Zeo
- -Some colonies are turning pinkish-red
- -Many colonies for diluted and non-diluted M13 genes+ZeoR on Zeo
- -Some colonies are turning pinkish-red
- -Many colonies for No DNA control on Zeo
- Could the white colonies be the designed colonies and can we kill the red colonies with Zeocin before killing the white colonies (aka. Use high Zeocin concentrations to select for the correct construct)
- -Selected 1 red colony from No DNA Zeo control plate and 1 white colony from M13genes+ZeoR sample plate
- -Added 100ul H2O then divided amongst 5 culture tubes each (with 5mL of low-salt LB, pH 7.5)
- -Then added Zeocin to a final concentration of: 0, 25, 50, 75, 100ug/mL.
- Results of 6/13 infection
- -Litmus 28i infected cells
- on Kan: Individual colonies for 1:10, 1:100, and 1:1000. No colonies on 1:5000
- on Amp: lawn for 1:10, 1:100, and near lawn for 1:1000. Single colonies for 1:5000
- -pSB1C3-M13ori infected cells
- on Kan: Same as Litmus 28i samples on Kan
- on Chlor: Lawn for all dilutions. Must discredit due to recent Chlor contamination
- -Litmus 28i infected cells
Week 7
6/15
- Results of Zeocin experiment on 6/14
- -Healthy growth for both at 0ug/mL Zeo
- -No growth for white colony with any Zeo
- -Good growth for red colony even at 100ug/mL Zeo
- -This suggests that the red colonies are naturally resistant to Zeocin. Also, our plates must not contain active Neocin. We are sure that we are adding enough. Possibly, we add it while the media is too hot or leave the plates at room temperature (decondensing) for too long and this deactivates the antibiotic? Perhaps we are not attaining the correct pH
- In light of recent contamination problems on both Chlor and Zeo, we made new competent cells
- -5alpha
- -BW23115
- -BW23115 conjugated (contains f-episome)
6/16
- Made new Chlor antibiotic. Made to Chlor plates
- Contamination Test: Streaked Zeo and new Chlor plates each with
- -Colony from Zeo contaminated plate
- -Colony from Chlor contaminated plate
- -Non-transformed OLD chemically competent cells
- -Non-transformed NEW chemically competent cells
- Redid bacterial infection
- -For both samples, we used ER2738 (not from the same ‘O/N’ (also, not a real O/N))
- -Infected one sample with isolated ‘Litmus28i phage’ and one with ‘pSB1C3-M13ori’
- -Alterations to protocol
- Did not started from saturated O/N. Started each with a colony, waited several hours until at OD ~1. We then added these cells to fresh 50mL LB to have an OD of 0.1.
- Missed the 30 minute infection mark. Infected for ~45 minutes.
- -Plated on Kan and Amp (Litmus28i sample) or Chlor (pSB1C3-M13ori sample) at dilutions of
- 1:100, 1:1k, 1:10k, and 1:100k
6/17
- Q5 PCR to replace KanR with ZeoR in M13K07
- -Used recommendations
- -Unfortunately, I (Jo) don’t know the difference between tightening and loosening the thermocycler lid; therefore, our M13K07 sample to amplify the M13K07 genes evaporated. But that’s ok, because we were sick of Zeocin anyway and decided mid-PCR to not waste our time with a Gibson Assembly and transformation. Instead we will be using the Old-School method of digestion and ligation because let’s face it, it’s a classic (and Gibson sucks) =)
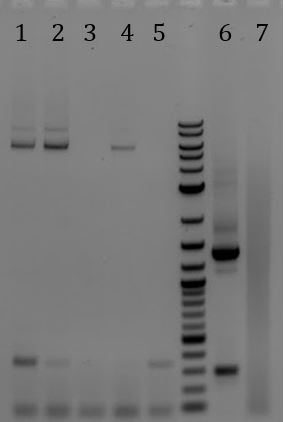
- Results from infection Test
| Litmus 28i | 1:100 | 1:1k | 1:10k | 1:100k |
|---|---|---|---|---|
| Amp | ~6000? | ~2056 | 377 | 36 |
| Kan | 130 | 8 | 3 | 0 |
| pSB1C3-M13ori | 1:100 | 1:1k | 1:10k | 1:100k |
|---|---|---|---|---|
| Chlor | 8 | 1 | 0 | 0 |
| Kan | 97 | 9 | 0 | 0 |
- -We consider the Litmus 28i to have been a success. The pSB1C3-M13ori …. not so much. Looking back at the Litmus28i and the M13K07, we noticed that the M13ori is facing the opposite direction as the plasmid ori. Ours faces the same direction as the plasmid ori. Because we are working with phagemids that are single stranded, we think that by flipping the M13ori, we may be able to recover functionality. We will also look into other reasons.
- Results from contamination test (6/16)
- -Old Chlor plate (6/12)
- -Non-transformed chemically competent cells
- Old: some colonies
- New: No colonies
- -Non-transformed chemically competent cells
- -New Chlor plate (6/16)
- -Colony from
- Zeo contaminated plate: No growth
- Chlor contaminated plate: Much growth
- -Non-transformed chemically competent cells
- Old: some colonies
- New: No growth
- -Colony from
- -Zeo (25ug/mL, Low NaCl, pH 7.5) (6/12)
- -Colony from
- Zeo contaminated plate: Much growth
- Chlor contaminated plate: No growth
- -Non-transformed chemically competent cells
- Old: some specks
- New: some specks
- -Colony from
- -Old Chlor plate (6/12)
6/18
- Ordered primers to…
- -biobrick M13ori (in other direction)
- -biobrick M13K07 genes
6/19
- Waited for primers
- Set up O/N cultures to test last infection (6/16) for colonies containing pSB1C3-M13ori
| Sample # | Selection | Dilution | Presumably | Notes |
|---|---|---|---|---|
| 1-8 | Chlor | 1:100 | pSB1C3-M13ori | |
| 9 | Chlor | 1:1k | pSB1C3-M13ori | |
| 10-14 | Chlor | 1:1k | pSB1C3-M13ori | These colonies were not present on plate during initial counting on 6/17 |
| 15-16 | Kan | 1:1k | M13K07 |
- -Selection and dilution refer to the plate. Cells were then grown under selection.
- -More colonies were seen on all Chlor plates. No new colonies appeared on Kan plates
- Tomorrow, we will mini-prep, digest, and run the samples on a gel to verify gene transfer.
6/20
- Primers are in!
- Cloning of pSB1C3-M13ori(New)
- -PCR to amplify M13ori (packaging signal) from Litmus 28i to be in the other direction
- -Primers (Gem011 F&R) add cut sites to make part biobrick compatible
- -Was able to gel extract
- Band at the same size as sample….. contaminated primers?
- -Then digested with E and P
- -Digested pSB1C3 plasmid with E and P to linearize backbone
- Was able to gel extract
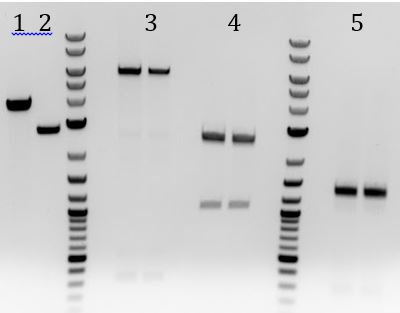
- -PCR to amplify M13ori (packaging signal) from Litmus 28i to be in the other direction
- 1. Spill over from 2
- 2 and 3. pSB1C3 digested with EcoRI and PstI
- -Insert <100bp so cannot be seen
- 4. M13ori
- Ligation of M13ori (packaging signal) into pSB1C3
- 10h at 16C
- 20m at 80C
- Hold at 4C
- PCR to amplify M13K07 genes from M13K07 DNA isolated from phage
- -Primers (Gem012 F&R) add cut sites to make part biobrick compatible
- -Very light bands and not band might correlate with 6000pb but not enough resolution on gel to be certain. Bands were too light so did not extract
- -Set up PCR again using Q5 to run O/N
- Received only a feint, smudgy band that was too large. We did not bother with extracting the DNA
- Test of 6/19 O/Ns (14 that are presumably pSB1C3-M13ori and 2 that are presumably M13K07)
- -Mini-prepped all O/Ns
- -Digested all with Pst-I (common RE between pSB1C3-M13ori and M13K07)
- -Even though these are phagemids, we assumed that because they were still in the cell, the plasmids were still double stranded so could be recognized by RE. Our assumption was valid
- -Results of digestion
- 1,2,4-14: all had bands that corresponded to pSB1C3-M13ori cut once
- 3: Slight band at expected size but very feint
- 15, 16: as expected, they contain bands of ~9000bp, correlating with the M13K07 phagemid. Interestingly, sample 15 also contained a (brighter) band that corresponds with pSB1C3-M13ori. We assume that this was an incident of double infection (chance or did this occur at high frequency?)
- Check table from 6/19 for more details
- -1-14: pSB1C3-M13ori
- -15-16: M13K07 from 1:1k diluted plate
- Check table from 6/19 for more details
6/21
- Needed to transform our pSB1C3-M13ori(New) (6/20) into cells. First, we transformed into 5alpha cells; however, we need to infect these cells later in order to make phage. Our 5alphas are not competent so we repeated the transformation, this time using ER2738 cells which contain the F’ episome, allowing us to infect with the M13 phage.
- -Also transformed sample #1 from 6/20 mini-preps (pSB1C3-M13ori(Old) into ER2738 cells. Do not see any notes about this plasmid being in infectable cells during the initial experiment. Reason for experiment failure???
- Ran 6/20 O/N PCR (to amplify M13K07 genes) on gel (also re-ran the previous sample from the day with more DNA)
- -Still, band looks too big. Brightest band still occurs between 8000 and 9000bp. Lighter band around 7000bp-- might be ~6000pb but too light to tell
- Alternative plan for amplifying M13K07 genes
- -Digest sample with PstI (cuts outside of the target region)
- Used sample #16 from 6/20 b/c it is double stranded so will cut
- -PCR amplified the linearized digestion product
- 1. PCR of M13genes
- 2. PCR of M13genes + DMSO
- Gel extracted pieces boxed in red
- -Used primers Gem012 F&R
- -Gel extracted piece but received very low yield
- Alternative to alternative plan for amplifying M13K07 genes
- -Digested with PstI and AgeI
- 1. Cut with PstI and AgeI
- 2. Uncut
- 3. Cut with PstI
- 4. Just PCR
- -Gel extracted ~6000pb band after double digestion
- -PCR (Phusion)
- -Digestion (A+P) product
- -Gel extraction product
- -No DNA control
Week 8
6/22
- Did receive colonies from 6/21 transformations. Selected colonies for overnight
| number | colonies |
|---|---|
| 1-7 | pSB1C3-M13ori(NEW) in 5alpha |
| 8-14 | pSB1C3-M13ori(NEW) in ER2738 |
| 15-18 | pSB1C3-M13ori(OLD) in ER2738 |
| 19 | p110+RBS in 5alpha as control |
| 20 | p110+RBS in ER2738 as control |
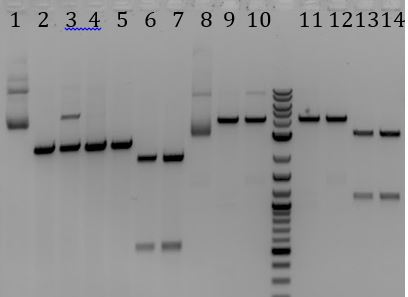
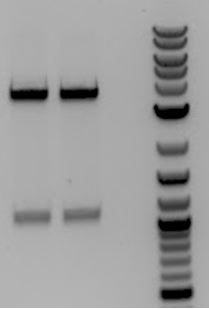
- Gel of 6/21 O/N PCR
- Digestion (AgeI+PstI) product ? Yielded the three bands that appear with every PCR of M13K07
- 1. M13K07-> digested (AgeI + PstI) -> PCR
- 2. M13K07-> digested (AgeI+ PstI) -> extraction-> PCR
- 3. No DNA control for PCR
- -Gel extraction product ? No bands
- -No DNA control ? No bands
- PCR purified the above PCR (Digestion (AgeI+PstI) product) and PCR from 6/21
- Digested PCR purified sample with EcoRI+PstI and ran gel
- 1. PCR-> PCR purified
- 2. Sample after DpnI digest
- 3. Sample after digestion with EcoRI and PstI
- 4. Samples after digestion with DpnI then EcoRI and PstI
- Ligation of above digestion (M13K07 genes) with pSB1C3
6/23
- Transformed pSB1C3-M13ori(New) (6/22 ligation)
- Check overnights from 6/22 for the correct insert
- -Mini-prep O/Ns
- -Digested with EcoRI and PstI
- 1-7: pSB1C3-M13ori(New) in 5alpha
- 8-14: pSB1C3-M13ori(New) in ER2738
- 15-18: pSB1C3-M13ori(Old)
- -All were the expected size (though gel wiggled)
- -Sent 4 samples for sequencing (iGEM primers: VF2 and VR)
- 4: Divergent (new) phagemid 1C3 transformed into 5alpha
- 11, 13: Divergent (new) phagemid 1C3 transformed into ER2738
- 16: Convergent (old) phagemid 1C3 transformed into ER2738
6/24
- Results of 6/23 transformation (pSB1C3-M13genes) from 6/22 ligation)
- -Received ~100 colonies
- -Set up O/N cultures for 8 of the colonies
- -Time passed…..
- -Mini-prepped the 8 O/N samples mentioned above (yes, it was a long day)
- Digested samples (E+P)
- Started phage amplification protocol
- -Amount of phage added = 10.9ul of 1:10 diluted phage (M13 phage (1) from 5/24 (4.575 x10^12 phage/mL)) to a final concentration of 1x10^8 phage/mL in 50mL
- -Managed to get both samples to the 14-28hr incubation
6/25
- Checked mini-prep samples from 6/24 (pSB1C3-M13genes)
- Ran the digestion overnight (6/24 to 6/25)
- -All contained a band at 2000bp. Mostly empty vector or contained small, light band. Sample 1 had prominent band at ~1250bp.
- -None of samples contained M13genes
- M13K07 is on P15A ori (10-12 copy number) whereas the pSB1C3 ori is on pUC19 (500-700 copies). It’s possible that this overexpression is detrimental to cell
- Alternative low copy plasmids found in distribution kit
- 2013 (plate 5)
- pSB6A1 (1K)
- pSB3C5 (3C)
- pSB3K3 (5E)
- 2014 (plate 4)
- pSB3C5 (4D)
- pSB6A1 (2L)
- -Suspended and transformed the above plasmids into 5alphas
- -Next, we will select colonies, mini-prep, digest, gel extract, ligate with M13genes
- Alternative low copy plasmids found in distribution kit
- Started phage amplification protocol
- -Phagemids in ER2738
- pSB1C3-M13ori(New)
- pSB1C3-M13ori(Old)
- -Set up O/N of ER2738 for infection tomorrow
- -Phagemids in ER2738
6/26
- Results of transformation of plasmids from the distribution kit
- -Only received colonies pSB6A1 (2L from 2014) and pSB3C5 (4D from 2014)
- -Pricked colonies for O/N
- PCR of M13 genes so we can ligate it into the above backbones tomorrow
- -DNA: M13K07 digested with EcoRI+PstI (6/21)
- -Primers: Gem012 F & R
- Finished phage amplification protocol
| Samples | A269 | A320 | Concentration |
|---|---|---|---|
| Old phagemid 1C3 (1) | 0.832 | 0.492 | 7.828 x10^12 |
| Old phagemid 1C3 (2) | 0.324 | 0.083 | 5.549 x10^12 |
| New phagemid 1C3 (1) | 0.391 | 0.077 | 7.229 x10^12 |
| New phagemid 1C3 (2) | 0.402 | 0.639 | -5.457 x10^12 |
- While resuspending “New phagemid 1C3 (2)”, the tip fell off and we lost half of sample. Evidently, we lost most of phage so we tossed sample
- -Prepped and Infected ER2738 with “Old phagemid 1C3 (1)” and “New phagemid 1C3 (2)”
- Added 1.6ul of 1:10 diluted “Old phagemid 1C3 (1) to ER2738 cells
- Added 1.8ul of 1:10 diluted “New phagemid 1C3 (1) to ER2738 cells
- -Plated the infected cells at dilutions
- 1:1 onto Chlor+Kan plates
- 1:10 onto Chlor+Kan plates
- 1:100 onto Chlor and onto Kan plates
- 1:1000 onto Chlor and onto Kan plates
- 1:10000 onto Chlor and onto Kan plates
- 1:100000 onto Chlor and onto Kan plates
6/27
- To ligate M13genes onto different backbones
- -Mini-prepped the O/Ns from 6/26 to get backbones with low copy number
- pSB6A1
- pSB3C5
- did not grow as well
- -PCR purified PCR (to amplify M13 genes with Gem012 F & R) from 6/26
- -Digestions (50ul)
- Digest pSB6A1 with E + P
- Digest pSB3C5 with E + P
- Digest PCR purification product with E + P
- -Mini-prepped the O/Ns from 6/26 to get backbones with low copy number
- 1. pSB6A1
- 2. pSB3C5
- 3. M13genes
- -Gel extraction of the above digestions
- For each: Added 10ul of 6x loading dye to 50ul digestions and divided the total volume between 2 wells
- -Ligation
- 1) pSB6A1-M13genes
- 2) pSB3C5-M13genes
- -Gel extraction of the above digestions
- Results from 6/26 infection (after 22hrs in incubator)
| Anti-Sense Phagemid 1C3 | 1:100 | 1:1000 | 1:10000 | 1:100000 |
|---|---|---|---|---|
| Chlor | 85 | 6 | 2 | 0 |
| Kan | 405 | 13 | 2 | 1 |
| Sense Phagemid1C3 | 1:100 | 1:1000 | 1:10000 | 1:100000 |
|---|---|---|---|---|
| Chlor | 121 | 24 | 3 | 0 |
| Kan | 25 | 1 | 0 | 0 |
-We also plated the infected cells on plates containing Chlor and Kan to test for the possibility of double infection
| Phagemid 1C3 | 1:1 | 1:10 |
|---|---|---|
| Anti-Sense | 59 | 0 |
| Sense | 4 | 0 |
6/28
- We noticed that there were more colonies on our infection plates from 6/26 than on 6/27; therefore, we recounted colonies
- -No increase of colonies on Kan plates
- -Significant increase of colonies on Chlor plates
- -Sense refers to the first phagemid 1C3 where the M13ori is in the sense direction compared to the plasmid ori
- -Anti-Sense refers to the new phagemid 1C3 where the M13ori is in the anti-sense direction compared to the plasmid ori
- Numbers on the left are after 22 hours. Numbers on the right are after 38.75 hours
- Results of transformation from 6/27 (pSB6A1-M13genes and pSB3C5-M13genes)
- -We have many colonies. Unfortunately, some are red, suggesting that original insert (J04450) was not successfully separated from backbone through gel extraction
- -Selected colonies to grow overnight in 5mL LB
Week 9
6/29
- Check O/N cultures for correct constructs (pSB6A1-M13genes and pSB3C5-M13genes)
- -Mini-prep samples
- -Digested with EcoRI and PstI to check insert sizes
- -Gel
- lanes….
- 1-11: pSB6A1-M13genes
- 12-18: pSB3C5-M13genes
- -Epic failure
- -All pSB6A1 backbones are empty
- -Half of pSB3C5 backbones were empty. The others contained random inserts (1700 OR 3500). We don’t know what these inserts are. 1700 band is likely the digestion product that appears when we digest M13genes
- lanes….
- Question: Can we use empty vectors from these mini-preps as ligation vectors?
- -Digest pSB6A1 mini-prep with…. (see if both cut sites were retained during re-ligation)
- no enzyme
- EcoRI
- PstI
- EcoRI + PstI
- -Digest pSB6A1 mini-prep with…. (see if both cut sites were retained during re-ligation)
- lanes....
- 1-2: last two samples of pSB3C5-M13genes from above gel
- 3. Uncut
- 4. Cut with EcoRI
- 5. Cut with PstI
- 6. Cut with EcoRI and PstI
- lanes....
- -Appears that two backbones are ligated together
6/30
- Made chemically competent ER2738 cells that contain Litmus28i DNA
- To amplify M13genes in order to retry ligation
- -PCR of M13K07 DNA (diluted 1:100) to amplify the M13 genes
- -PCR purify PCR product
- -Run on gel: PCR purification, PCR, noDNAcontrol
- PCR and purification showed bands at ~9k; therefore, did not get product
- No DNA control was clean
- Made freeze downs of
- pSB3C5-J04450
- pSB6A1-J04450
- ‘empty’ pSB3C5
- ‘empty’ pSB6A1
7/1
- Digestion of PCR product from 6/30 to figure out where mistake is
- -Not really useful
- Lanes...
- 1. PCR pur -> digested with AgeI
- 2. PCR pur -> digested with NgoMIV
- 3. PCR pur -> digested with PstI
- 4. PCR pur -> uncut
- 5. PCR pur -> digested with dpnI
- 6. Uncut plasmid DNA
- Talked to Mary: She says we were adding to much DNA
- -PCR again. Used 1:100 dilution of 1:100 diluted M13K07 DNA. (aka 1:10000 dilution)
- Received a beautiful band at 6k bp
- lanes
- 1. 1:100 dilution
- 2. 1:1000 dilution
- 3. 1:10 000 dilution
- 4. No DNA control
7/2
- To biobrick M13 genes (pSB6A1-M13genes and pSB3C5-M13genes)
- -PCR purified 7/1 PCR product (Primers = Gem012)
- -Digested with EcoRI-HF and PstI-HF
- -Ligation to
- pSBA61 (digested and gel extracted)
- pSB3C5 (digested and gel extracted)
7/3
- Transformation
- -pSB6A1-M13genes into 5alpha cells and ER2738 with pSB1C3-M13ori
- -pSB3C5-M13genes into 5alpha cells and ER2738 with Litmus28i
- -Transformed into cells containing a phagemid in order to skip some steps
7/3-7/7 Vacation!
Week 10
7/7
- Made O/N cultures of 7/3 transformation colonies
- -Transformation results were not recorded until 7/8 (see below)
7/8
- Transformation results ( CC = Chemically comp cells, ER = ER2738)
- -No growth on No DNA controls
- ER-Litmus28i CC on AMP-Chlor-Tet
- ER-phagemid1C3 CC on AMP-Chlor-Tet
- 5alpha CC on Amp
- 5alpha CC on Chlor
- -Lots of red colonies on positive controls, no white colonies
- ER-Litmus28i CC + pSB3C5 on AMP-Chlor-Tet
- ER-phagemid1C3 CC + pSB6A1 on AMP-Chlor-Tet
- -Lots of white colonies on sample plates, some red colonies
- ER-Lit CC + M13genes-pSB3C5 on AMP-Chlor-Tet
- ER-phagemid1C3 CC + M13genes-pSB6A1 on AMP-Chlor-Tet
- 5alpha CC + M13genes-pSB3C5 on Chlor
- 5alpha CC + M13genes-pSB6A1 on Amp
- -No growth on No DNA controls
- Overnights from 7/7 look healthy
- -Renamed O/N to have numbers instead of long names
| Sample # | Cells | DNA |
|---|---|---|
| 1-5 | ER-Lit | M13genes-pSB3C5 |
| 6-10 | ER-phagemid1C3 | M13genes-pSB6A1 |
| 11-15 | 5alpha | M13genes-pSB3C5 |
| 16-20 | 5alpha | M13genes-pSB6A1 |
- -Mini-prepped DNA
- -Digest the mini-preps with EcoRI-HF and PstI-HF
- -Run digestions on gel to check sizes
- lanes
- top-left: pSB3C5-M13genes in ER2738 with Litmus28i
- top-right: pSB6A1-M13genes in ER2738 with pSB1C3-1C3
- bottom-left: pSB3C5-M13genes in 5alpha
- bottom-right: pSB6A1-M13genes in 5alpha
- None are correct
- Primers Gem013 came in. Resuspend and diluted primers
- -O/N of pSB3C5 to use for PCR tomorrow
7/9
- Wanted to make Litmus28i biobrick compatible for use as a phagemid backbone for us and other iGEM teams
- -Mini-prepped O/N of pSB3C5
- -PCR of pSB3C5 to amplify J04450 with Litmus28i compatible cut sites
- Primers: Gem013
- At the 3’ end, these primers are the same as VF2 and VR so will bind the region flanking J04450. This conserves the terminators that exist between the biobrick prefix and VF2 on one side and those between the biobrick suffix and VR on the other side. Therefore, we are amplifying, VF2 priming site, terminators, J04450, terminators, and VR priming site
- At the 5’ end, these primers contain unique restriction sites found in the Litmus28i MCS
- Primers: Gem013
- PCR of M13 genes…. again (did 4 samples)
- -Used the 1:10000 dilutions
7/10
- Note: The following two projects were done in parallel when possible
- To make Litmus28i biobrick compatible
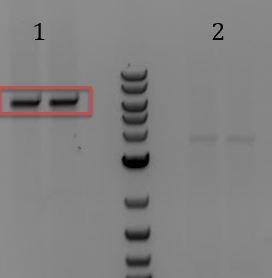
- -Ran gel of PCR from 7/9 (J04450 amplification with Gem013)
- 1-4: PCRs of M13genes
- 5. No DNA control for M13genes PCR
- 6. PCR of pSB3C5 backbone
- 7. No DNA control for pSB3C5 backbone
- -PRC purified the PCR product
- -Digestion #1
- Restriction enzymes had the same buffer conditions but different activation temperatures so we had to do a 2 part digestion
| Sample | Restriction enzymes | Notes |
|---|---|---|
| J04450 | Sac1 | PCR purified |
| Litmus 28i | Sac1 | From NEB tube |
- Incubate 1hr at 37 C
- Heat inactivated 20 minutes at 80C
- -Digestion #2
- -Added 1ul BsmI to both samples
- -Incubated 1hr at 65C
- -Heat inactivated 20 minutes at 80C
- Ran gel
- -See gel below
- -Tried to extract J04450 segment but received very low yield. Since band otherwise looked clean, we decided to redo digestion then skip straight to ligation
- Repeat digestion #1 for J04450
- Repeat digestion #2 for J04450
- Ligation (10hr at 16C, 10 min at 80C)
- 3. Litmus28i + J04450
- To retry ligation to biobrick backbone
- -Ran gel of PCR from 7/9 (M13genes amplified with Gem012)
- See gel below
- -PCR purified the PCR product
- -Digestion
- -Ran gel of PCR from 7/9 (M13genes amplified with Gem012)
| Sample | Restriction enzymes | Notes |
|---|---|---|
| M13 genes | EcoRI-HF + PstI-HF | PCR purified |
| 6A1 | EcoRI-HF + PstI-HF | ‘empty’ pSB6A1 |
| 3C5 | EcoRI-HF + PstI-HF | contains J04450 |
- Incubate 1hr at 37C
- Heat inactivated 20 minutes at 80C
- -Ran gel
- See gel below
- Tried to extract M13genes and pSB3C5 segments but received very low yields. Since bands otherwise looked clean, we decided to redo digestion then skip straight to ligation
- -Repeat digestion for M13genes and pSB3C5
- -Ligation (10h at 16C, 10m at 80C)
- 1. pSB3C5 + M13genes
- 2. pSB6A1 + M13genes
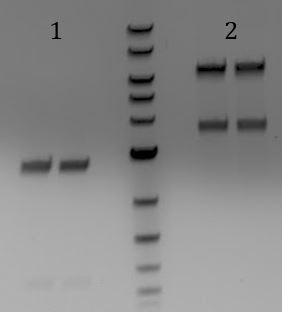
- Gel
- 1. pSB1A3
- 2. Limtus28i
- 3. M13genes
- 4. pSB3C5
- 5. J04450
7/11
- Transformed ligations from 7/10 into 5alpha cells
- 1. pSB3C5 + M13genes
- 2. pSB6A1 + M13genes
- 3. Litmus28i + J04450
7/12
- Results of 7/11 transformation
- -No growth on no DNA control (Amp or Chlor)
- -Lots of growth on Lit-J04450 -> some red-> colonies are too close together to prick individual colonies
- -It’s possible that these red colonies are satellites =(
- -Swiped some and plated on new Amp plate (restreak)
- -Lawn of positive control (Litmus 28i) on Amp
- -Many colonies for L1 (pSB3C5-M13genes) and L2 (pSB6A1-M13genes)
- -Due to high number of red colonies on Lit-J04450 plate, we assumed that most of these colonies contain empty vector
- -Did not make O/N
- Ligated M13 genes to pSB6A1
- -Used the remained of our digested M13genes.
- -Both digestions from 7/10
- Infection Experiment
- -had 3 cell stocks (each taken from a different colony the night before)
- -Tested Litmus 28i, Tandem phagemid 1C3, and double infection
- Therefore, there were 9 flasks total.
- -negative control: Streaked parent cells (non-infected) onto Chlor, Amp, and kan
- -Included double infection plates for Litmus 28i and phagemid 1C3
Week 11
7/13
- Transformed 7/12 ligation (pSB6A1-M13genes)
- Litmus28i-J04450
- -All growth from 7/12 restreak was white
- -Pricked some red colonies for liquid O/N-will hopefully see red tomorrow
- -Pricked a few red colonies and put into 200ul H2O (a few colonies per tube- 2 tubes total).
- Plated 150ul onto Amp plates
- -Put original plate into incubator to hopefully get bigger colonies
7/14
- Finished TWIV ppt
- Set up 2 liquid culture of red Litmus 28i colonies (Litmus28i-J04450)-- slow growth
- -Mini-prepped red Litmus 28i ‘O/N’ from earlier in day
- -Digested samples—to check for insert and correct cut sites
- Made O/Ns of pSB6A1-M13genes colonies from 7/13 transformation
7/15
- Ran gel of Litmus28i-J04450 samples
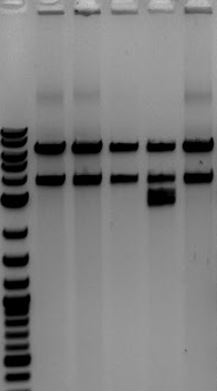
- 1. J04450-Litmus28ibb #1 (EcoRI+PstI)
- 2. J04450-Litmus28ibb #1 (uncut)
- 3. J04450-Litmus28ibb #2 (EcoRI+PstI)
- 4. J04450-Litmus28ibb #2 (EcoRI+PstI)
- Accidentally added restriction enzymes
- 5. Litmus28i (EcoRI+PstI)
- Only has PstI site
- -Verifies that Litmus28i-J00450 had correct cut sites
- -From this point on, biobricked Litmus28i is called Litmus28ibb
- Check 7/14 O/Ns for pSB6A1-M13genes
- -Mini-prepped liquid cultures
- -Digested with EcoRI and PstI
- Received bold bands of just under 4000bp. Some lanes had a very feint band ~330bp. None were the correct size
- Transformed Litmus28ibb-J04450 into ER2738 cells
- Remake phage packaging
- -pSB1C3-M13ori(New)
- -pSB1C3-M13ori(Old)
7/16
- Results of 7/15 transformation of Litmus28i into 6/30 ER2738 cells
- -lawn on no DNA control-- most likely contaminated cells
- Streaked chem comp 5alpha and ER onto Amp and Amp+Tet plates to determine if problem is with cells or plates
- -O/N culture of ER to make new chem comp cells
- -lawn on no DNA control-- most likely contaminated cells
7/17
- Finished phage isolation
- Note:
- pSB1C3-M13ori (New): M13ori and plasmid ori are convergent
- pSB1C3-M13ori (Old): M13ori and plasmid ori are tandem
- Note:
| 269nm | 320nm | [] phage/mL | |
|---|---|---|---|
| pSB1C3-M13ori (New) | 0.386 | 0.034 | 8.126 E12 |
| pSB1C3-M13ori (New) | 0.419 | 0.036 | 8.842 E12 |
| pSB1C3-M13ori (Old) | 0.283 | 0.025 | 5.956 E12 |
| pSB1C3-M13ori (Old) | 0.326 | 0.024 | 6.972 E12 |
- Infection to compare the new and the old pSB1C3-M13ori
- -Prepped cells for infection
- -Infected cells with pSB1C3-M13ori (New) or pSB1C3-M13ori (New)
- -and plated on dilutions of 1:100, 1:1000, and 1:10000 on Chlor and kan
- -Also plated 1:1000 dilution on lowered Chlor concentration (34ug/mL)
- Sent Litmus28ibb-J04450 for sequencing
7/18
- Results from contamination test
- -5alpha on Amp = no growth
- -5alpha on Amp+Tet = colonies
- -ER on Amp = lawn
- -ER on Amp+Tet = lawn
- Made new chemically competent ER2738
- Transformed Litmus28ibb-J04450 into ER2738 made on 3/30 and ER2738 made on 7/18
- -Had two samples of Litmus28ibb-J04450 and two cell stocks, so 4 samples total
7/19
- Results of 7/17 infection
- -pSB1C3-M13ori(Old) (tandem-when plasmid ori and M13ori point in the same direction) packages better than pSB1C3-M13ori(New) (when ori and M13ori are convergent), implying that directionality matters.
Week 12
7/20
- Freeze downs
- Litmus28i-J04450 Litmus 28i is now biobrick compatible
7/21
- Send samples for sequencing
- pSB1C3-M13ori(Old)
- pSB1C3-M13ori(New)
- To ligate M13ori to a kanamycin backbone (pSB1K3)
- Resuspended pSB1K3-J04450 from distribution kit (6B on plate 4)
- Contains J04450 as insert (full RFP construct)
- Transform pSB1K3 into 5alpha cells
- Resuspended pSB1K3-J04450 from distribution kit (6B on plate 4)
7/22
- Pricked colony from pSB1K3-J04450 transformation
- Mini-prepped DNA to get a supply of DNA
- To put M13ori (M13 phackaging signal) onto Kanamycin resistance o we can test packaging ratios with M13K07 on the same antibiotic
- Digested pSB1K3 with EcoRI-HF and XbaI
- Digested pSB1C3-M13ori (Old) with EcoRI-HF and SpeI-HF
- DNA was not sufficiently cut. Too much DNA? Problem with enzyme (SpeI-HF)?
- 1. pSB1K3
- 2. M13ori
7/23
- Sequencing samples from 7/21 were lost in the mail. Resent samples
- Yet another attempt to biobrick M13genes
- 1. using primers that would amplify genes and M13 ori parts from M13K07
- 2. using primers that would amplify genes, M13 ori parts, and plasmid ori from M13K07
- 1. Amplified only the M13genes and M13ori
- 2. No DNA control for 1
- 3. Amplified M13genes, M13ori, and plasmid ori
- 4. No DNA control for 3
- When run on a gel, samples were clean with only one band at around 7kb. No contamination in no DNA controls
- Digestions
- pSB1K3 with EcoRI-HF and PstI-HF to check for correct insert
- pSB1C3-M13ori (Old and new) with EcoRI-HF and SpeI-HF to test for efficient cutting with different stock of restriction enzyme
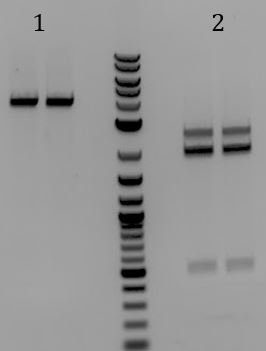
- 1-3: pSB1K3
- 4. pSB1C3-M13ori (Old)
- 5. pSB1C3-M13ori (New)
- Still had inefficient cutting. Tested M13ori next to Litmus28i to disern the problem
- Uncut
- Cut once with (E, X, S, or P)
- Cut twice with (E+S or E+P)
- 1. M13ori uncut
- 2. M13ori Ecori-HF
- 3. M13ori SpeI-HF
- 4. M13ori XbaI
- 5. M13ori PstI-HF
- 6. M13ori EcoRI-HF + SpeI-HF
- 7. M13ori EcoRI-HF + PstI-HF
- 8. Litmus28i uncut
- 9. Litmus28i EcoRI-HF
- 10. Litmus28i SpeI-HF
- 11. Litmus28i XbaI
- 12. Litmus28i PstI-HF
- 13. Litmus28i EcoRI-HF + SpeI-HF
- 14. Litmus28i EcoRI-HF + PstI-HF
- Only partial digest with only SpeI-HF for both. Complete digestion with all others, including E+S
7/24
- To Biobrick M13genes using pSB6A1
- PCR purified M13 genes (did both samples at the same time)
- Digested PCR purification and pSB6A1 with EcoRI-HF and PstI-HF
- Gel extracted M13genes
- 1. M13genes
- 2. pSB6A1
- Bands of pSB6A1 were too light to gel extract
- To swap the kanamycin resistance marker on M13K07 with ampicillin resistance
- PCR amplified AmpR from pSB6A1
- made O/N culture of pSB6A1 from freeze down to mini-prep
- Also streaked cells onto plate
7/25
- To change resistance marker on M13K07
- 1. Biobrick method
- Tried to mini-prep O/N but pellet (after liquid culture was spun) was not red even after 16+ hours. Set up O/N from plate colonies
- 2. Swap only resistance method
- PCR purified 7/24 PCR
- Ran product on gel-> band of correct size
- 1. Biobrick method
- 1. PCR product
- 2. PCR product-> PCR purified
- 3. No DNA control
- Digested the PCR purification products of M13genes+ori and AmpR with AgeI-HF and NotI-HF
- Bands on previous gel looked clean enough for both so did not gel extract
- Overnight ligation
- Set up O/Ns of pSB1K3 colonies
7/26
- Transform
- pSB6A1+M13genes
- To biobrick M13genes
- Digestion
- pSB6A1 with EcoRI and PstI
- Digestion
- Gel extracted backbone (band ~4000bp)
- Ligations (10hrs 16C, 10min at 80C)
- pSB6A1+M13genes
Week 13
7/27
- Transformation results from 7/26
- No growth for no DNA controls **No growth for M13 genes onto pSB6A1 *Set up O/Ns of 5alpha and ER2738 to make competent cell tomorrow
- Made freeze down of pSB1K3
- Plated ER competent cells from 5/15 and 6/16 Onto Chlor plates to check for contamination
7/28
- No growth of ER2738 on Chlor (either sample)
- ER is not contaminated with ChlorR
- Make new 5alpha chem comp cells
- Transformations
- pSB6A1-M13genes
- positive control for Amp (pSB6A1)
- No DNA control
7/29
- Set up O/Ns of pSB6A1-M13genes
- Plated sample on reduced Amp and regular Tet
- Later in day…
- Mini-prepped O/Ns
- Digested and ran on a gel to check for insert sizes
-
- All but the 4th lane with 3 bands look correct
-
7/30
- Lawn on last night’s plating-> replate
- Digest pSB3C5 and pSB6A1-M13genes with E and P to move M13genes to a Chlor backbone
- Extracted bands
- Ligations
- pSB3C5 + M13genes
- Sent samples for sequencing
- M13 genes on pSB6A1 VF2
- M13 genes on pSB6A1 VR
Test experiment for high school kids coming to lab (7/29-7/31)
- General idea: Have 2 strands of DNA, one has an EcoRI site while the other contains a SNP, abolishing the cut sight. We pretend that one is pathogenic (the one that is not cut) and tells kids to figure out which one is which
- PCR with Dream-Taq
- Did not have special fastdigest enzyme
- Hypothesized that green dye will interfere with enzyme effectivity
- PCR with Dream-Taq (no green dye)
- Digest with EcoRI fastdigest-> did not cut
- Redid experiment several times
- Should digest with XbaI
- PCR purified PCR products
7/31
- Spent the morning with Heritage High School
- Made phage
| Phage | A269 | A320 | genome size | [ ] |
|---|---|---|---|---|
| Litmus28ibb-J04450 (1) | 0.823 | 0.197 | 9.35 x10^12 | |
| Litmus28ibb-J04450 (2) | 0.940 | 0.168 | 1.14 x10^13 |
8/2
- Digestion
- pSB6A1-M13genes**pSB3C5
Week 14
8/3
- Another attempt to clone M13genes onto pSB3C5
- Run 8/2 digestions on gel
- 1. M13genes (E+P)
- 2. pSB3C5 (E+P)
- Gel extracted
- M13genes away from pSB6A1 backbone
- pSB3C5 backbone away from J04450
- Ligations
- pSB3C5 + M13genes
- pSB3C5 + no insert
- Transformed ligations into ER2738
8/5
- Transformation of pSB3C5+M13genes failed
- Made phage
- Fd-CAT DNA packaged with Fd-CAT
- phagemid 1C3 packaged with M13K07
- amilCP on pSB1C3 packaged with M13K07
- Litmus28ibb-J04450 packaged with M13K07
8/6
- Isolated phage
| Phage | [ ] |
|---|---|
| Fd CAT | 2.72 x10^12 |
| Litmus28ibb-J04450 | 8.37 x10^12 |
| pSB1C3-amilCP | 1.42 x10^13 |
| pSB1C3-M13ori | 4.32 x10^12 |
8/6
- Started making phage with Fd CAT the should contain Litmus28ibb-RFP
- Infection
- Infected ER2738 cells with Litmus28ibb-RFP, pSB1C3-M13ori, or pSB1C3-amilCP
- Plated Litmus samples on 100ug/mL Amp and 50ug/mL Kan
- Plated other two samples on 34ug/mL Chlor and 25ug/mL Kan
8/7
- Isolated phage containing Litmus28ibb-RFP using helper phage
- M13K07 (as control)
- Fd-CAT (no phage pellet was observed-worried that there was no phage)
- Went to CSU to have them test M13ori part compared to amilCP. Grew the samples in 5mL O/Ns then diluted to 0.5OD and grew 30 minutes rather than starting from a fresh colony
8/8
- At CSU
- Finished phage protocol (Test packaging of M13ori part)
- Only used 20mL infection samples
- grew to 0.55 OD
- When making phage (after 14 hour incubation), there was very little growth
- Plated non-diluted and diluted 1:1000 of M13ori and amilCP sample
- Infected cells using phage isolated 8/7. Plated at dilutions of 1:100 and 1:100k
- Want to test progeny Fd-CAT phage (made 8/7) for infectability
- Start phage isolation prodocol using Fd-CAT phage from 8/7 do deliver the helper phagemid
- Phagemid:
- None: will make Fd-CAT phage packaing Fd-CAT phagemid
- Litmus28ibb-J04450: make Fd-CAT phage packaging Litmus28ibb-J04450 phagemid
- Start phage isolation prodocol using Fd-CAT phage from 8/7 do deliver the helper phagemid
- Remake phage (Fd-CAT) using fresh stock from Mike
8/9
- Results of 8/8 infection
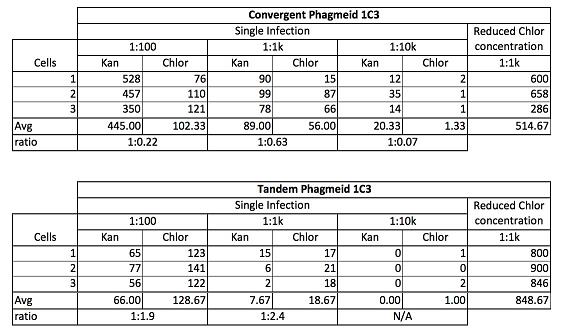
| Helper Phagemid | phagemid | Selection | 1:1000 dilution | 1:00k dilution |
|---|---|---|---|---|
| M13K07 | Litmus28ibb-RFP | Kan+Tet | 400-500 (some red) | 61 white; 20 red |
| same | same | Amp+Tet | lawn | 2 |
| Fd-CAT | Litmus28ibb-RFP | Chlor+Tet | 0 | 0 |
| same | same | Amp+Tet | 10 | 707 |
- NOTE: The same sample was plated on two different plates. For example, the sample using M13K07 as the helper phage was plated on Kan+Tet and Amp+Tet
- Mini-prep
- pSC3C5-J04450 -- very little growth, low DNA yield
- Fd-CAT infected cells
- Digest and run on a gel to verify presence of a band- there was a very feint band
- Finished isolating phage…. messed up and used 0.8MgCl2, 0.2M NaCl instead of PEG during precipitation step. Also used the wrong phage
- Infected ER2738 cells anyway…. no growth by 8/12
Week 15
8/10
- Are the progeny phage from 8/6 (original progeny from first Fd-Tet application) viable/ able to reproduce?
- Used these progeny to make phage that should amplify Fd-CAT phage containing either the Fd-CAT or Litmus28ibb-J04450 phagemid
8/11
- Finished isolating phage
| Helper Phagemid | Phagemid | A269 | A320 | genome size | [ ] |
|---|---|---|---|---|---|
| Fd-CAT | Litmus28ibb (1) | 0.029 | 0.013 | 4080 | 2.35 x10^11 |
| Fd-CAT | Ltimus28ibb (2) | 0.033 | 0.015 | 4080 | 2.65 x10^11 |
| Fd-CAT | Fd-CAT (1) | 0.326 | 0.082 | 7775 | 1.88 x10^12 |
| Fd-CAT | Fd-CAT (2) | 0.320 | 0.079 | 7775 | 1.86 x10^12 |
| M13g6A1 | M13g6A1 (1) | 0.322 | 0.148 | 10,029 | 1.04 x10^12 |
| M13g6A1 | M13g6A1 (2) | 0.503 | 0.130 | 10,029 | 2.23 x10^12 |
8/13
- Infected ER2738 with Fd-CAT packaging Litmus28ibb-J04450
8/16
- Results from Infection on 8/11
| Helper Phagemid | Phagemid | 1:1 | 1:10 | 1:100 | 1:1000 |
|---|---|---|---|---|---|
| M13g6A1 | M13g6A1 | almost lawn | 728 | 287 | 55 |
| Fd-CAT | Fd-CAT | lawn | almost lawn | 1260 | 640 |
| Fd-CAT | Litmus28ibb-J04450 | lawn | almost lawn | 2004 | 304 |
 "
"