Team:CU-Boulder/Notebook/CC9Phage Team
From 2014.igem.org
Note:
Litmus28ibb is a biobricked version of the Litmus28i (from NEB) vector which contains the M13ori (packaging signal) which allows for uptake into M13 or Fd phage. While making this part biobrick compatible, we added J04450 as the insert.
pCas9 (BBa_K1218011) contains and tracrRNA and Cas9 gene under native promoters, and a minimal CRISPR arrays. The guide RNA (referred here as gRNA) is inserted into the CRISPR array.
Contents |
Week 1
7/21
- To ligate pCas9 to Litmus28ibb
- -Digestion
- -Litmus28ibb-J04450 with E+P
- -pCas9 with E+P
- -Digestion
- 1. pCas9
- 2. Litmus28ibb
- -Overnight ligations (10hrs at 37C, 10min at 80C)
7/22
- Transformed ligation from 7/21 (Litmus28ibb + pCas9) into ER2738 cells
- -Plated on Amp + Tet
- -Later in day, pricked colonies from morning transformation (Litmus28ibb and pCas9 in ER)
7/23
- Miniprepped O/N from 7/22 of Litmus28ibb-pCas9
- -Digested
- None were the correct size. Smaller bands looked like pSB1C3. Plated cells onto Chlor and there was significant growth. pSB1C3 from the pCas9?
- -Digested
7/24
- To put pCas9 onto Litmus28i
- -Digested
- -Litmus28ibb with EcoRI+PstI
- -pSB1C3-pCas9 with EcoRI+SpeI
- -PSB3C5-J04450 with XbaI + PstI
- -Digested
- 1. Litmus28ibb
- 2. pCas9
- 3. J04450
- -Overnight ligation (10hr @ 16C)
7/25
- Transformed ligation from 7/24 (Litmus28ibb+pCas9) into 5alpha cells
7/26
- No growth from 7/25 transformation
- Tranform
- -Litmus28ibb + pCas9 + J04450
- Retry making Litmus28i with pCas9
- -Ligation (10hrs at 16C, 10min at 80C)
- Litmus28ibb + pCas9 + J04450
- -Ligation (10hrs at 16C, 10min at 80C)
Week 2
7/27
- Transformation results
- -No growth for no DNA controls (on Chlor)
- -4 colonies for Litmus28bb-JO4450-Cas9 (on Amp-> none are red)
- Picked all 4 colonies for O/Ns
7/28
- 3 new red colonies on Litmus28bb-Cas9-J04450 plate
- -Set up cultures
- O/Ns grew but did not bother
- Transformations
- -Litmus28bb-J04450-Cas9
- -No DNA control
7/29
- Litmus28bb-Cas9-J04450 O/Ns
- -Incubator shut off during night. Samplers were at RT by morning
- -Add 1mL to fresh 4mL LB and Amp
- -Mini-prep DNA
- -Digest with EcoRI and PstI to check insert for correct size
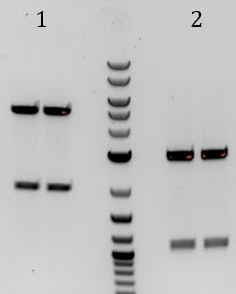
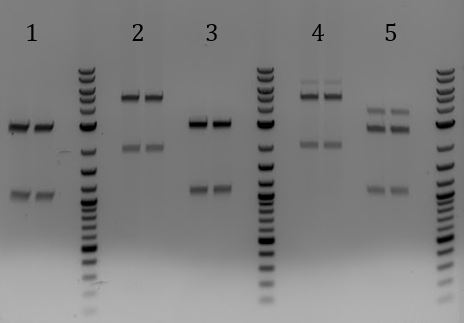
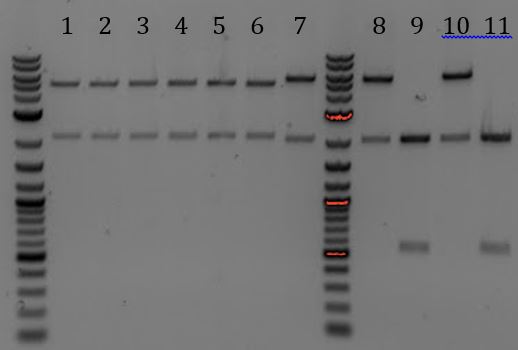
- -Lane 2 contains the expected 3000bp band for the Litmus28ibb backbone and the ~7000bp band for the pCas9-J04450 insert. Unfortunately, this sample also contains a 4000bp band.
- Plated the above sample
7/30
- Digested Litmus28ibb-Cas9-RFP with EcoRI-HF
- -Extracted large band ~9000
- -Re-ligated piece
- Sent samples for sequencing
- -Litmus28bb with Cas9 and RFP (LCR) gRNA sequence
8/1
- Mini-prepped O/Ns of
- -Litmus28ibb-Cas9-J04450 (LCR)
- Digested with EcoRI-HF and PstI-HF => all samples had band ~3700
- -Litmus28ibb-Cas9-J04450 (LCR)
- Digestions of Litmus and Cas9 to retry ligation
- 1. Litmus28ibb (E+P)
- 2. pCas9(E+P)
- 3. Litmus28ibb (E+P)
- 4. pCas9(E+S)
- 5. pSB3C5 (X+P)
- -Gel extractions
- -Ligations
- Litmus28ibb (E+P) to pCas9 (E+P)
- Litmus28ibb(E+P) to pCas9(E+S) and J04450(E+P)
8/2
- Transformation into BW cells (5/30)
- -Hundreds of colonies for LCR Miniprep
- Digestion to get a better idea of what this DNA is
8/3
- Isolate the LCR from the extra band in the gel
- -Run 8/2 digestions on gel
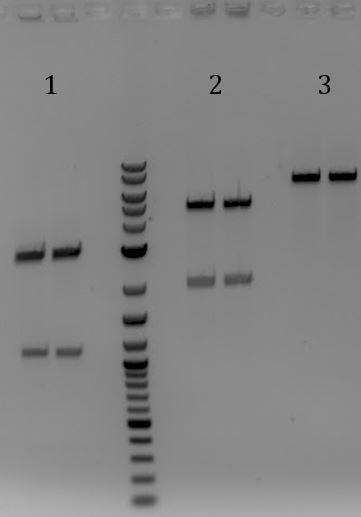
- 1. LCR (PstI)
- 2. LCR (EcoRI)
- -Gel extracted
- -LCR digested with PstI-HF (top band
- -Re-ligate extracted LCR with no insert (should re-ligate to itself)
- -Transformed ligation into ER2738
- -Gel extracted
- Made O/Ns
- -colonies from 8/2 transformation
- -restreak of LCR from freeze down
Week 3
8/4
- Check overnights from 8/3 (LCR = Litmus28ibb+pCas9+RFP) for correct inserts
- -Mini-prep samples
- -Digest with EcoRI and PstI
- All had sizes ~3700bp
- No correct inserts
- Attempt to ligate Litmus28ibb and pCas9(gRNA3)
- -Digestions
- Litmus28ibb-RFP E+P extracted Lit back bone
- pCas9 gRNA3 E+P extracted pCas9 insert
- -Digestions
- 1. Litmus28ibb
- 2. pCas9(gRNA3)
- 3. pCas9(unmod.)
- -Ligations (10 hrs at 16C, 10 min at 80C)
- Litmus28ibb + pCas9(gRNA3)
- -Ligations (10 hrs at 16C, 10 min at 80C)
8/5
- Transformation into ER2738 (7/18), 5ul of ligation products
- 1. Litmus28ibb-Cas9gRNA3
8/6
- Another attempt at ligating pCas9 and Litmus28ibb
- -Digest unmodified pCas9 with EcoRI-HF and PstI-HF
- -Digest pCas9 containing pRNA1 with EcoRI-HF and PstI-HF
- 1. pCas9(unmod.)
- 2. pCas9(gRNA1)
- -Gel extract pieces
- -Ligate separately to Litmus28ibb
- Results of 8/5 transformation
- -6 colonies for Litmus28ibb+pCas9(gRNA3) on Tet+reduced Amp
- Picked colonies for overnights
- -No growth for no DNA control
- -6 colonies for Litmus28ibb+pCas9(gRNA3) on Tet+reduced Amp
- Transformation of ligations from earlier today
- -Litmus28ibb + pSB1C3-pCas9(unmod.)
- -Litmus28ibb + pSB1C3-pCas9(gRNA1)
- Mini-prep Litmus28ibb+pCas9(gRNA3) O/N samples from earlier in day
- -Digested with EcoRI-HF and PstI-HF
- All samples had bands of 3000 and 2000bp. None were correct
- -Digested with EcoRI-HF and PstI-HF
8/8
- M13ori into SB Cas9
- -Digestions
- -unmodified pCas9 with EcoRI-HF and XbaI
- -M13ori with EcoRI-HF and SpeI-HF
- -pCas9(gRNA1) with EcoRI-HF and XbaI
- -pCas9(gRNA3) with EcoRI-HF and XbaI
- -Digestions
8/9
- To clone M13ori onto pSB1C3-pCas9
- -Run 8/8 digestion on a gel
- 1. M13ori
- 2. pCas9(gRNA3)
- 3. pCas9(gRNA1)
- -Gel extracted
- -Ligation Ligated M13ori to….
- -pSB1C3-pCas9(gRNA1)
- -pSB1C3-pCas9(gRNA3)
- -pSB1C3-pCas9(unmod.)
- -Transform and plate onto low Chlor 934 ug/mL)
- -Ligation Ligated M13ori to….
Week 4
8/10
- Digested ligation products from 8/6 and 8/4. Saw many large bands that may have been undigested
- O/Ns of 8/9 transformation
- -1 colony for M13ori-pCas9(gRNA3) (only 1 colony grew)
- -11 colonies for M13ori-pCas9(gRNA1)
- -No colonies grew for M13ori-pCas9(unmodified)
8/11
- Test 8/10 O/Ns for correct inserts
- -Digest 8/10 O/Ns with EcoRI-HF and PstI-HF
- 1. M13ori-pCas9(gRNA3)
- 2-12. M13ori-pCas9(gRNA1)
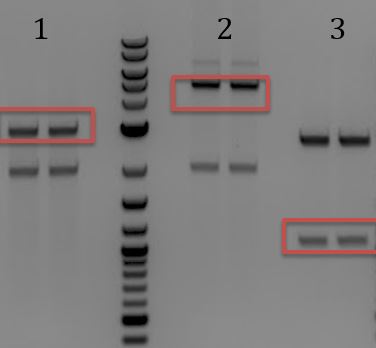
- Lanes 1, 8, and 9 are about the right size. Sent these samples for sequencing
- To ligate M13ori to pSB1C3-pCas9(unmod.)
- -Digest
- pSB1C3-pCas9(unmod.) (E+P)
- M13 digested 8/8
- -Ligation: See L5 below
- -Digest
- To ligate pCas9 and M13ori to pSB1A3
- -Digest
- pSB1A3 (E+P)
- M13ori (E+S)
- Unmodified pCas9 (X+P)
- pCas9(gRNA1)-M13ori (E+P)
- pCas9(gRNA3)-M13ori (E+P)
- -Ligation: See L2-4 below
- -Digest
- Another try to put pCas9 onto Litmus28ibb
- -Digested
- Unmodified Litmus28i (E+P)
- pCas9(unmod.) (E+P)
- pCas9(gRNA1)-M13ori (E+P)
- pCas9(gRNA3)-M13ori (E+P)
- -Ligation: See L1 and L6 below
- -Digested
- Gel for all
- 1. Unmodified Litmus28i
- 2. pSB1C3-pCas9(unmodified)
- 3. pSB1A3
- 4. pSB1C3-M13ori
- 5. pSB1C3-pCas9(unmod.)
- 6. pSB1C3-M13ori-pCas9(gRNA1)
- 7. pSB1C3-M13ori-pCas9(gRNA3)
- Ligations for both projects
| # | Backbone | Insert 1 | Insert 2 |
|---|---|---|---|
| 1 | Unmod. Litmus28i (E+P) | pCas9(unmod.) (E+P) | N/A |
| 2 | pSB1A3 (E+P) | pCas9(unmod.) (X+P) | M13ori (E+S) |
| 3 | pSB1A3 (E+P) | pCas9(gRNA1)-M13ori (E+P) | N/A |
| 4 | pSB1A3 (E+P) | pCas9(gRNA3)-M13ori (E+P) | N/A |
| 5 | pSB1C3-pCas9(unmod.) (E+X) | M13ori (E+S) | N/A |
| 6 | Litmus28ibb (E+P) | pCas9(gRNA3) (E+P) | N/A |
| 7 | Litmus28ibb (E+P) | pCas9(gRNA1) (E+P) | N/A |
| 8 | Litmus28ibb (E+P) | pCas9(unmod.) (E+P) | N/A |
8/12
- Transform ligation from 8/11 into ER2738 cells
- Make phage using M13g6A1 as Helper Phage
- -used pCas9(gRNA1)-M13ori or pCas9(gRNA3)-M13ori as phagemid
- Freeze downs
| Top | Side |
|---|---|
| M. gRNA3 Cas9 (8/12) | M13ori Cas9 gRNA3 pSB1C3 |
| M. gRNA1 Cas9 (8/12) | M13ori(8) Cas9 gRNA1 pSB1C3 |
| M. gRNA1 Cas9 (8/12) | M13ori (9) Cas9 gRNA1 pSB1C3 |
8/13
- Finish phage isolation
| Helper phage | Phage | A269 | A320 | genome size | [ ] |
|---|---|---|---|---|---|
| Fd-CAT | M. gRNA1 | 0.353 | 0.025 | 7687 | 2.56 x10^12 |
| Fd-CAT | M. gRNA3 | 0.554 | 0.037 | 7687 | 4.40 x10^12 |
- Infection
- -Infecting ER2738 and BWF’
- -Phage
- pSB1C3-pCas9(gRNA1)
- pSB1C3-pCas9(gRNA3)
- Transformation Results from 8/12
- -Growth for the following… (made O/Ns)
- pSB1A3-M13ori-pCas9(gRNA3)
- pSB1C3-M13ori-pCas9(unmod)-M13ori
- Litmus28ibb-pCas9(gRNA3)
- Litmus28ibb-pCas9(gRNA1)
- Litmus28ibb-pCas9(unmod.)
- -Hundreds of colonies for pCas9(unmod.) on Unmod. Litmus28i. Didn’t trust result so didn’t pick coloneis. EcoRI and PstI sites touch so most likely, Litmus28i was only cut once
- -Growth for the following… (made O/Ns)
8/15
- Check O/Ns from 8/14 for correct inserts
- -Mini-prepped samples
- -Digested with E and P
- -Results
- No bands of the right size for
- -Litmus28ibb-pCas9(gRNA3)
- -Litmus28ibb-pCas9(gRNA1)
- -Litmus28ibb-pCas9(unmod.)
- -Expected band sizes for… (see below)
- pCas9(gRNA3)-M13ori on pSB1A3
- pCas9(unmod)-M13ori on pSB1C3
- Transform into BWF’ to test for functionality of Cas9 when associated with m13ori, independent of infectivity
- -M13ori-pCas9(gRNA1)
- -M13ori-pCas9(gRNA3)
8/16
- PCR on LCR (Litmus28ibb with pCas9 and RFP to maybe get a good product to religate
- -Didn’t work
Results of 8/13 infection of BWF’ with M13ori-gRNA(1 or 3) to test pCas9 killing
- No growth on DNA controls on Chlor(34ug/mL) or Kan(50ug/mL)+Chlor(34ug/mL)
- ON Kan(50ug/mL) + Chlor (170ug/mL) in BWF'
- gRNA1--> 22 colonies
- gRNA3--> 2 colonies
On Kan(25ug/mL)+Chlor(34ug/mL) in BWF’
| 1:100 | 1:1000 | |
|---|---|---|
| gRNA1 | 617 | 84 |
| gRNA3 | 469 | 42 |
On Chlor (34ug/mL) in BWF’
| 1:100 | 1:1000 | |
|---|---|---|
| gRNA1 | 508 | 67 |
| gRNA3 | 447 | 41 |
On Chlor (34ug/mL) in ER
| 1:100 | 1:1000 | |
|---|---|---|
| gRNA1 | 702 | 93 |
| gRNA3 | 518 | 58 |
Results of 8/15 transformation of BWF’ with M13ori—gRNA(1 or 3) to test killing
Chlor (34ug/mL) + Kan (50ug/mL)
| 1:1 | 1:10 | |
|---|---|---|
| gRNA1 | Lawn | 1904 |
| gNRA3 | Slightly thinner lanw | 1616 |
Chlor (34 ug/mL)
| Trial 1 | Trial 2 | |
|---|---|---|
| gRNA1 | 1788 | 1548 |
| gRNA3 | 1564 | 1416 |
Samples were diluted 1:10
Week 5
8/17 Make phage
- pSB1C3-M13ori-pCas9(unmod)
- pSB1A3-M13ori-pCas9(gRNA3)
Ran uncut LCR on a gel and gel extracted the three bands
8/18 While making phage…. We threw away the samples…. Remake phage. Same as 8/17 Transform LCR gel extractions into 5alpha cells
8/19 Sent samples for sequencing
- pSB1C3-M13ori-pCas9(unmodified)
- pSB1A3-M13ori-pCas9(gRNA3)
Finish making phage
| Name | A269 | A320 | Genome Size | [ ] |
|---|---|---|---|---|
| pSB1A3-M13ori-pCas9(gRNA3) | 0.697 | 0.162 | Note: We later found that this sample did not actually contain the M13ori | |
| pSB1C3-M13ori-pCas9(unmod) | 0.764 | 0.146 | 7687 | 3.044 x10^13 |
Infect cells (ER and BWF’)
- Grew a single 75mL sample. When OD ~0.5, divided samples into 3
- Added 20mL of sample to 125mL flask (3 for each strain)
- Added phage to a final concentration of 1 x10^8 phage/mL
- Incubated at 250rpm, 37C, for 30 minutes
- Plated at dilutions (1:10, 1:100, and 1:1k)
- On Chlor(170ug/mL), Kan(50ug/mL)+Chlor(85ug/mL), Kan(50ug/mL)+Chlor(170ug/mL)
8/21
Finished phage amplification protocol from 8/20 but did not receive any phage.
Infect BWF’ cells with the phage having pSB1C3-M13ori-pCas9 containing the scramble, gRNA1, and gRNA3.
'8/22 Present project to department
Week 6
8/25 Results of 8/21 infection experiment
Results on Kan and Chlor
| Scramble | gRNA1 | gRNA3 | |
|---|---|---|---|
| 1:1 | 149 | 143 | 13 |
| 1:10 | 14 | 16 | 1 |
Results on Chlor
| Scramble | gRNA1 | gRNA3 | |
|---|---|---|---|
| 1:1 | 147 | 117 | 14 |
| 1:10 | 14 | NA | 3 |
8/28 Retry ligation of pCas9 to litmus28ibb Digest Litmus28ibb-J04450, pCas9, and amilCP with EcoRI and PstI. Gel extract pieces Ligate pCas9 into Litmus28ibb and amilCP into Limus28ibb
8/29 Transform 8/28 ligation products into 5alpha cells
8/29 Transform pSB1C3-pCas9(unmodified) into BW and BWF’ to test targeting of f’ episome. Due to poor infection results we wanted to test the guide RNAs for targeting of the F’ episome. We have searched the vast webs for the sequence of the F’ episome but have yet to (and never will) find it.
8/30 Transformation results of pCas9 into BW and BWF’. Results were unexpected, we expected more growth for BW sample or at least equal growth. Differences in competence or concentration could explain results in initial count
| Sample | Initial count | After another 24 hours |
|---|---|---|
| No DNA controls (for BW and BWF’) | No growth | No growth |
| BW cells | 9 colonies | 222 |
| BWF’ cells | ~100 colonies | 158 |
Transformation results for ligation of pCas9 to Litmus28ibb
| Amp (50ug/mL) | Amp (100ug/mL) | Notes | |
|---|---|---|---|
| No DNA control | 3 | 10 | All bright white |
| Litmus28ibb-pCas9 | ~50 (many are red) | 25-30 | High proportion are red. Some are shadowy white. A few are bright white |
| Litmus28ibb-amilCP | >300 | >300 | Mostly shadowy, some red, some bright white |
Made overnight cultures of shadowy and bright coloneis from Litmus28ibb-pCas9 samples (from both plates) and of shadowy colonies from Litmus28ibb-amilCP. Since growth on no DAN control was bright white, we assumed that bright colonies on sample plates were more likely to be contamination, but didn’t really know. Since there were fewer colonies on no DNA control plates, we figures we could randomly select colonies from the sample plates that were not a result of contamination.
Week 7
8/31 Mini-prep overnight cultures from 8/30 and digest samples with EcoRI and PstI. When run on a gel, none of the Litmus28ibb-pCas9 samples yielded bands of the correct size. All Litmus28ibb-amilCP samples did yield the expected band sizes.
9/2 Sent some of mini-prep samples from 8/31 for sequencing
9/3 Digestion of pCas9 with EcoRI and speI and the unmodified NEB version of Litmus28i with EcoRI and XbaI. Gel extracted pieces and ligated together (No, this part would not have the BioBrick prefix/suffix)
9/4 Transform 9/3 Litmus28i-pCas9 ligation into 5alpha cells. Selected on low ampicillin (50ug/mL) and high ampicillin (100ug/mL)
9/5 Did receive colonies from 9/4 transformation. Selected 24 colonies for colony PCR with a primer pair that would span the junction of pCas9 and Litmus28i. (forward primer bound to CRISPR region of pCas9 while reverse primer bound in the middle of the Litmus28i backbone) Colony PCR failed to reveal any promising samples. Set up overnight cultures in a hopeless effort to yield different results.
Week 8
9/7 Mini-prepped overnight cultures from 9/5. Digested each with EcoRI and PstI and ran reults on gel. Results were consistent with colony PCR: failure. A few samples had very feint bands that were approximately the correct size
9/8 for the samples on 9/7 with light bands of approximately the correct size, we sent samples for sequencing (another hopeless attempt to get different results)
9/9 Sequencing results showed that Limtus28i religated
Week 9
9/17
New method to clone pCas9 onto Litmus28ibb vector: PCR->Digestion->ligation method
- PCR amplified pCas9 construct from pSB1C3-pCas9. PCR amplified Litmus28ibb backbone from Litmus28ibb-J04550
- PCR purified both samples. Digested both samples with EcoRI and PstI and gel extracted the pieces
- Ligated pCas9 to Litmus28ibb vector
Sent pSB1C3-M13ori-pCas9 sample for sequencing to sequence the entire plasmid
9/18
Transform 9/17 ligation (Litmus28ibb-pCas9) into 10beta cells
9/19
There was growth on no DNA control. Some colonies for sample.
9/20
Make chemically competent ER and 5alpha cells using DMSO from bottle instead of fancy tubes. We think this is the reason for later transformation failures
Digested pCas9 and Litmus28ibb PCR purifications from 9/17 with EcoRI, PstI, and DpnI. PCR purified digestion samples then ligated pieces together.
Week 10
9/21
New idea: Cloning pCas9 onto Litmus28i has not worked up until this point. What if we constructed a vector that looked similar to litmus28i. pSB1A3 has the same origin of replication and almost the same ampicillin resistance marker (with a few SNPs). Need to add M13ori (which was taken from Litmus28i) and pCas9. M13ori would not be in the correct position on plasmid but this is as close as we can get with the remaining time
- Digestions: M13ori-pCas9(unmod), pCas9(unmod), pSB1A3 with EcoRI and PstI. Gel showed incomplete digestion so did not gel extract
Transformation into 5alpha cells: no DNA control, 9/20 ligation (Litmus28ibb-pCas9), pSB1C3-amilCP, and pSB1C3-M13ori-pCas9
9/22
Re-digested M13ori-pCas9(unmod) and pSB1A3-J04450 with EcoRI and PstI. pCas9(unmod) was PCR purified (product looked clean on gel so did not bother to gel extract). Ligated M13ori-pCas9(unmod) to pSB1A3 and pCas9(unmod) to pSB1A3 Sent pSB1A3 to M13ori-pCas9 for sequencing to cover region not covered by previous sequencing
9/23
Sequencing results came back
- Litmus28ibb-J04450 contains two nucleotides that are flipped. They are located within the ampicillin resistance gene and do change two amino acids; however, sample grows on ampicillin so these are not loss of function mutation.
- pSB1C3-M13ori-pCas9 was perfect - the above two samples are ready to submit to the registry
Transform ligations from 9/22 (pSB1A3- M13ori-pCas9(unmod) and pCas9(unmod)) into 5 alpha cells Colony PCR 18 samples from 9/20 Limtus28ibb-pCas9(unmod) plate.
9/24
Run gel of 9/23 colony PCR to check for desired Litmus28ibb-pCas9(unmod). There was a band of approximately the correct size in each samples (including the no DNA control). Set up overnight cultures of first 5 samples for further testing Results of 9/23 transformation
- No growth for no DNA control on chlor (170ug/mL), Amp (50ug/mL) or Amp (100ug/mL)
- 7 colonies for pSB1C3-M13genes on chlor (170ug/mL)
- 27 colonies for pSB1A3-pCas9 on Amp (100ug/mL) and 41 colonies on Amp (50ug/mL)
- 0 colonies for pSB1A3-M13ori-pCas9 on Amp(50 and 100ug/mL)
Set up overnight cultures of some of the samples.
9/25
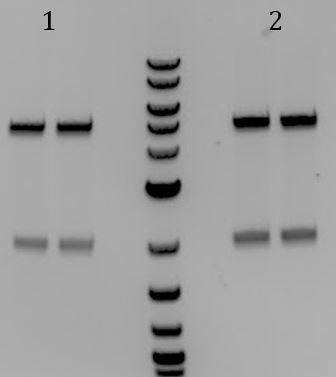
Mini-prep overnights from 9/24 Digest samples to check for insert. When run on a gel, 3 samples had large bands of ~5000bp and one had an additional band at ~6000. We made overnights of these 3 colonies
9/26
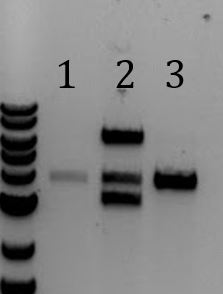
Digested mini-prepped DNA from the three samples that looked promising from 9/25. Each sample was digested with only EcoRI, only PstI, or EcoRI and PstI. When run on a gel, it can be seen that each samples, when cut once, yielded one band 8000bp while when cut twice, yielded 2 bands of ~2000 and 5000 as expected. Sent parts to iGEM registry: pSB1C3-M13ori-pCas9 and Litmus28ibb-J04450 To add the M13ori to pSB1A3-Cas9
- Digested pSB1A3-pCas9 with EcoRI and XbaL
- Digested pSB1C3-M13ori with EcoRI and SpeI
- Gel extracted bands of the correct sizes and combined these into a ligation reaction.
9/27 Transform pSB1A3-M13ori-pCas9 into Amp(50ug/mL) and Amp(100ug/mL) Later in the day…. Set up overnight cultures of colonies
Week 11
9/28
Mini-prepped overnights made 9/27 Digested overnight samples with EcoRI and PstI Did receive pSB1A3-M13ori-pCas9 samples with bands of the correct size (as compared to pSB1A3-pCas9) Set up overnight cultures of the samples that looked promising Transformed pSB1A3-M13ori-pCas9(unmod) and pSB1A3-M13ori-pCas9(gRNA3) into ER2738 cells
9/29
Did receive colonies from 9/28 transformation. Set up overnight cultures for both. Also set up overnight cultures of ER2738 and BWF’ for upcoming infection. Start phage amplification protocol using pSB1A3-M13ori-pCas9(unmod) and pSB1A3-M13ori-pCas9(gRNA3) as the phagemid. Used Fd-Cat as the helper phagemid
9/30
Finished phage amplification/isolation protocol for Fd phage packaging pSB1A3-M13ori-pCas9(unmod) and pSB1A3-M13ori-pCas9(gRNA3)
10/1
Measured concentration of phage isolated on 9/30. There were no phage for the gRNA3 sample. The pSB1A3-M13ori-pCas9(unmod) sample contained some phage but had a high level of contamination/impurity. Send samples pSB1A3-M13ori-pCas9(unmod) and pSB1A3-M13ori-pCas9(gRNA3) samples for sequencing to verify the presence of the M13ori
10/2
Sequencing results came in
- pSB1A3-M13ori-pCas9(unmod) looks good
- pSB1A3-M13ori-pCas9(gRNA3) doesn’t have the M13ori
Plan
- 1.SDM on pSB-M13ori-pCas9(unmod) to remove BsaI cut site in pSB1A3 backbone
- 2.Change spacer from unmodified to the gRNA3
- 3.Redo the infection experiment
SDM
- PCR to amplify pSB1A3-M13ori-pCas9(unmod)
- Treat with kinase, Ligase, and DpnI
- Transform into NEB 5alpha cells
10/3
Set up O/N cultures of 10/2 transformation
10/4
Mini-prep overnights. Samples are called pSB1A3(delta BasI)-M13ori-pCas9(unmod) to indicate each part of the construct and the removed BsaI site in the pSB1A3 backbone. Changing the gRNA spacer in pCas9
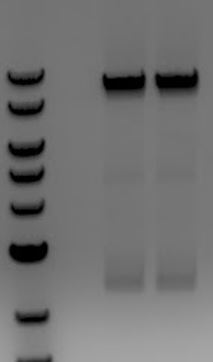
- Vector digest: digested pSB1A3(delta BasI)-M13ori-pCas9(unmod) with BsaI to remove unmodified spacer. Saw only one band on the gel (cut around spacer) which shows that the additional BsaI site in the pSB1A3 backbone was removed. Gel extracted the digested vector
- Ligation: Used the annealed oligos for gRNA3 (our targeting sample) from the last time we changed the spacer. Ligated these oligos to the digested pSB1A3(delta BasI)--M13ori-pCas9
Week 12
10/5
Transform pSB1A3(delta BasI)-M13ori-pCas9(unmod) and pSB1A3(delta BasI)-M13ori-pCas9(gRNA3) in ER2738 cells
10/6
Transformation results from 10/5
- No growth for no DNA control
- pSB1A3(delta BasI)-M13ori-pCas9(unmod) = many colonies
- pSB1A3(delta BasI)-M13ori-pCas9(gRNA3) = no colonies
Re-anneal primers then relegate
- Phosphorylation of oligos for gRNA3. Incubate for 1 hour at 37C then 20 minutes at 65C
- Add 2.5ul 1M NaCl
- Anneal in thermocycler. 5 minutes at 95C then a slow cool-down for ~2 hours
- Dilute product 1:10
- Ligation of newly annealed primers to digest and gel extracted pSB1A3(delta BasI)-M13ori-pCas9(unmod) from 10/5
10/7
Transform 10/6 ligation into ER2738 cells
10/8
No growth for 10/7 transformation Transform ligation from 10/6 into 5alpha cells Last try to make pSB1A3-M13ori-pCas9(gRNA3)
- Know from 10/1 sequencing that we have pSB1A3-pCas9(gRNA3). Therefore, only need to add M13ori upstream of pCas9 part
- Digest pSB1A3-pCas9(gRNA3) with EcoRI and XbaI and pSB1C3-M13ori with ExoRI and SpeI
- Gel extract digested pieces
- Ligate gel extracted pieces together
10/9
No growth for 10/8 transformation Transform 10/8 ligation into 5alpha cells
10/10
Transform
- pSB1C3-M13ori-pCas9(unmod) from 10/9 into ER2738 cells
- pSB1A3 (delta BsaI)-M13ori-pCas9(gRNA3) from 10/6 into 5 alpha cells
- pSB1A3-M13ori-pCas9(gRNA3) from 10/8 into 5 alpha cells
10/11
Transformation results of 10/10
- 1 colony for pSB1C3-M13ori-pCas9(unmod) in ER2738 (does not seem trustworthy)
- No colonies for no DNA control
- No colonies for the 2 other transformations
Transform pSB1C3-M13ori-pCas9(unmod) into ER2738 cells again Start phage amplification protocol to make the following phage
| Phagemid | Helper Phagemid |
|---|---|
| pSB1C3-M13ori-pCas9(unmod) | M13g6A1 |
| pSB1C3-M13ori-pCas9(unmod) | M13K07 |
| pSB1C3-M13ori-pCas9(gRNA3) | M13g6A1 |
| pSB1C3-M13ori-pCas9(gRNA3) | M13K07 |
pSB1C3-M13ori-pCas9(unmod) grew unexpectedly fast so we missed the OD mark. Growth rate compared to pSB1C3-M13ori-pCas9(gRNA3) sample suggests contamination. Did not proceed with making these phage for today.
10/12
No growth for 10/11 transformation – conclude that ER2738 cells are not competent (these are the cells made 9/20 with DMSO from a room temp bottle instead of the alliquotes from NEB. Remake phage
| Phagemid | Helper Phagemid |
|---|---|
| pSB1C3-M13ori-pCas9(unmod) | M13g6A1 |
| pSB1C3-M13ori-pCas9(unmod) | M13K07 |
10/13
Finish phage isolation protocol and measure
| Phagemid | Helper Phagemid | Dilution | A269 | A320 | Genome size | Diluted [ ] | Actual [ ] |
|---|---|---|---|---|---|---|---|
| pSB1C3-M13ori-pCas9(gRNA3) | M13g6A1 | 1:40 | 0.090 | 0.015 | 7687 | 5.85 E11 | 2.34 E13 |
| pSB1C3-M13ori-pCas9(gRNA3) | M13K07 | 1:40 | 0.085 | 0.013 | 7687 | 5.62 E11 | 2.25 E13 |
| pSB1C3-M13ori-pCas9(unmod) | M13g6A1 | 1:40 | 0.104 | 0.022 | 7687 | 6.40 E11 | 2.56 E13 |
| pSB1C3-M13ori-pCas9(unmod) | M13K07 | 1:20 | 0.091 | 0.016 | 7687 | 5.85 E11 | 1.17 E13 |
Infection with phage of BW23115F’ cells
- Grew a single stock of cells then dived into 4 samples each with 50mL cells. Added the above phage
- Incubated for 30 minutes at 37C, shaking
- Plated onto Chlor
Week 13
10/14
Infection results
- No growth on Chlor (170ul/mL)
- Very little to no growth on pCas9(unmod) samples. We think this is b/c the colony picked on 10/11 did not actually contain pSB1C3-M13ori-pCas9(unmod) so nothing was packaged into phage
- ~100 colonies for pCas9(gRNA3) samples.
- The samples made with M13g6A1 were also plated on ampicillin. These plates show considerable growth. There is more growth on
pCas9(unmod) than pCas9(gRNA3) sample plate. We believe this further supports our hypothesis that our pCas9(unmod) sample did not contain pSB1C3-M13ori-pCas9(unmod) so packaged M13g6A1 in the absence of a better phagemid. Also, for the pCas9(gRNA3) sample, there was considerably more growth on the ampicillin plate than on the chlor plate. This could be because pSB1C3-M13ori-pCas9(gRNA3) successfully killed the BW23115F’ cells as desired or the phagemid was not effectively packaged
Transformed pSB1C3-M13ori-pCas9(unmod) into chemically competent ER2738 cells that we know work from July.
 "
"