Team:CU-Boulder/Biobricks
From 2014.igem.org
| (33 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{: | + | {{Template:UCB-Main}} |
{{Template:UCB-NavBar}} | {{Template:UCB-NavBar}} | ||
| - | Part Summaries | + | == '''Part Summaries''' == |
| + | |||
For detailed characterization, see the parts page. | For detailed characterization, see the parts page. | ||
| - | + | <br> | |
| - | ==Part: BBa_K1445000 (M13ori)== | + | <br> |
| + | |||
| + | ===Part: BBa_K1445000 (M13ori)=== | ||
| + | |||
The M13 origin of replication (M13ori) has been documented as the packaging signal for the M13 phage. When single stranded, the M13ori forms unique hairpins that signal packaging into a phage capsid. It was experimentally demonstrated that this part is both necessary and sufficient for plasmid packaging into M13 phage. This part does not code for phage coat proteins; therefore, cannot produce phage. | The M13 origin of replication (M13ori) has been documented as the packaging signal for the M13 phage. When single stranded, the M13ori forms unique hairpins that signal packaging into a phage capsid. It was experimentally demonstrated that this part is both necessary and sufficient for plasmid packaging into M13 phage. This part does not code for phage coat proteins; therefore, cannot produce phage. | ||
| + | <br> | ||
| + | ===Part: BBa_K1445001 (Endogenous Type II CRISPR-Cas9 phagemid)=== | ||
| - | |||
This composite part consists of parts BBa_K1445000 (M13ori) and BBa_K1218011 (cas9). | This composite part consists of parts BBa_K1445000 (M13ori) and BBa_K1218011 (cas9). | ||
The cas9 construct includes the native trcrRNA and promoter region upstream of the type II Cas9 endonuclease. Downstream of the Cas9 protein is a minimal CRISPR array that contains the spacer region that determines the DNA sequence that is targeted by the Cas9 endonuclease. The spacer sequence can be easily replaced through a digestion-ligation reaction using BsaI. The addition of the M13 origin of replication (M13ori) allows for the uptake of the CRISPR-Cas9 machinery into M13 phage. | The cas9 construct includes the native trcrRNA and promoter region upstream of the type II Cas9 endonuclease. Downstream of the Cas9 protein is a minimal CRISPR array that contains the spacer region that determines the DNA sequence that is targeted by the Cas9 endonuclease. The spacer sequence can be easily replaced through a digestion-ligation reaction using BsaI. The addition of the M13 origin of replication (M13ori) allows for the uptake of the CRISPR-Cas9 machinery into M13 phage. | ||
| Line 19: | Line 24: | ||
| - | ==Part: BBa_K1445002 (High copy BioBricked M13 phagemid vector)== | + | ===Part: BBa_K1445002 (High copy BioBricked M13 phagemid vector)=== |
| + | |||
This biobrick backbone has the high copy number pUC19 origin of replication and an ampicillin resistance marker. It also contains the M13 origin of replication which allows for the phagemid to be packaged into the M13 phage. It was experimentally validated that vector retains its ability to be packaged into M13 phage. | This biobrick backbone has the high copy number pUC19 origin of replication and an ampicillin resistance marker. It also contains the M13 origin of replication which allows for the phagemid to be packaged into the M13 phage. It was experimentally validated that vector retains its ability to be packaged into M13 phage. | ||
'''[[File:UCB-Biobrick-02-141016.JPG]]''' | '''[[File:UCB-Biobrick-02-141016.JPG]]''' | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | {{Template:UCB-Footer}} | ||
Latest revision as of 23:09, 17 October 2014
Contents |
Part Summaries
For detailed characterization, see the parts page.
Part: BBa_K1445000 (M13ori)
The M13 origin of replication (M13ori) has been documented as the packaging signal for the M13 phage. When single stranded, the M13ori forms unique hairpins that signal packaging into a phage capsid. It was experimentally demonstrated that this part is both necessary and sufficient for plasmid packaging into M13 phage. This part does not code for phage coat proteins; therefore, cannot produce phage.
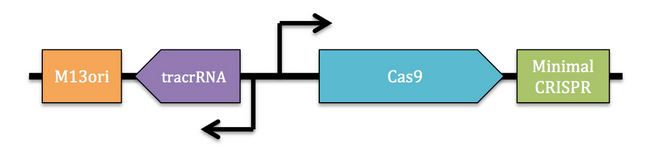
Part: BBa_K1445001 (Endogenous Type II CRISPR-Cas9 phagemid)
This composite part consists of parts BBa_K1445000 (M13ori) and BBa_K1218011 (cas9). The cas9 construct includes the native trcrRNA and promoter region upstream of the type II Cas9 endonuclease. Downstream of the Cas9 protein is a minimal CRISPR array that contains the spacer region that determines the DNA sequence that is targeted by the Cas9 endonuclease. The spacer sequence can be easily replaced through a digestion-ligation reaction using BsaI. The addition of the M13 origin of replication (M13ori) allows for the uptake of the CRISPR-Cas9 machinery into M13 phage.
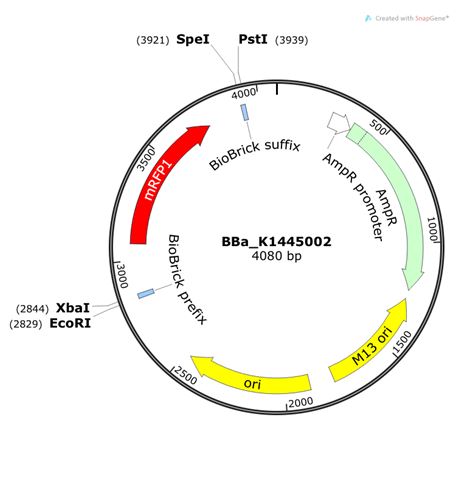
Part: BBa_K1445002 (High copy BioBricked M13 phagemid vector)
This biobrick backbone has the high copy number pUC19 origin of replication and an ampicillin resistance marker. It also contains the M13 origin of replication which allows for the phagemid to be packaged into the M13 phage. It was experimentally validated that vector retains its ability to be packaged into M13 phage.
 "
"